

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.5 + phase: 0 /pseudo
(550 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
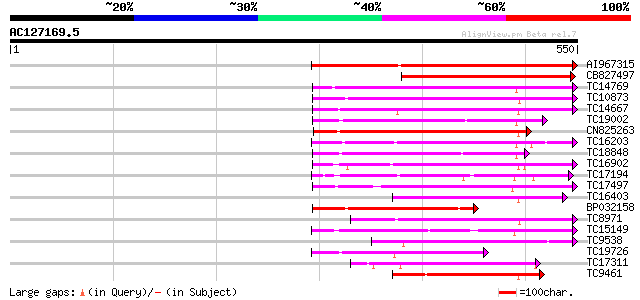
Score E
Sequences producing significant alignments: (bits) Value
AI967315 342 1e-94
CB827497 224 4e-59
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 188 2e-48
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 186 6e-48
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 182 2e-46
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 178 2e-45
CN825263 173 6e-44
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 172 1e-43
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 172 1e-43
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 159 1e-39
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 157 5e-39
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 135 2e-32
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 133 6e-32
BP032158 133 8e-32
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 133 8e-32
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 126 1e-29
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 123 7e-29
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 120 6e-28
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 119 1e-27
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 116 8e-27
>AI967315
Length = 1308
Score = 342 bits (876), Expect = 1e-94
Identities = 156/258 (60%), Positives = 198/258 (76%)
Frame = +1
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GGY+EVYKG L +G+ IAVKRL + +D KEKEFL E+G IGHVCH N LLG C +
Sbjct: 172 KGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEIGTIGHVCHSNVMPLLGCCID 351
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
NGLYL+F S G++++ +H A LDW RYKI +G ARGLHYLHK C+ RIIHRD
Sbjct: 352 NGLYLVFELSTVGSVASLIHDEKMA--PLDWKTRYKIVLGTARGLHYLHKGCQRRIIHRD 525
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASN+LL D+EPQI+DFGLAKWLP++WTHH++ P+EGTFG+LAPE +MHG+VDEKTD+
Sbjct: 526 IKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTFGHLAPEYYMHGVVDEKTDV 705
Query: 473 FAFGVLLLEIVTGRRPVDSSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLT 532
FAFGV LLE+++GR+PVD S Q++ WAKP++ I +L DPR+EG YDV Q RV
Sbjct: 706 FAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKWEIEKLVDPRLEGCYDVTQFNRVAFA 885
Query: 533 ASYCVRQTAIWRPAMTEV 550
AS C+R ++ WRP M+EV
Sbjct: 886 ASLCIRASSTWRPTMSEV 939
>CB827497
Length = 550
Score = 224 bits (570), Expect = 4e-59
Identities = 103/169 (60%), Positives = 132/169 (77%)
Frame = +2
Query: 381 LDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNK 440
L W IR+ IA+G ARGL YLH+ C+ RIIHRDIKA+N+LL DYEPQI DFGLAKWLP
Sbjct: 8 LPWCIRHNIALGTARGLLYLHEGCQRRIIHRDIKAANILLTEDYEPQICDFGLAKWLPEN 187
Query: 441 WTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDSSKQNILLWA 500
WTHH V EGTFGYLAPE +HGIVDEKTD+FAFGVLLLE+V+GRR +D S+Q+++LWA
Sbjct: 188 WTHHTVAKFEGTFGYLAPEYLLHGIVDEKTDVFAFGVLLLELVSGRRALDFSQQSLVLWA 367
Query: 501 KPLMESGNIAELADPRMEGRYDVEQLYRVVLTASYCVRQTAIWRPAMTE 549
KPL++ I EL DP + +D Q+ ++L AS C++Q++I RP++ +
Sbjct: 368 KPLLKKNEIMELIDPSLADGFDSRQMNLMLLAASLCIQQSSIRRPSIRQ 514
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 188 bits (478), Expect = 2e-48
Identities = 98/262 (37%), Positives = 152/262 (57%), Gaps = 5/262 (1%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ VY+G L +G+ +AVK + + +EF EL ++ + H N LLGYC E+
Sbjct: 2190 GGFGSVYRGTLNDGQEVAVK--VRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNES 2363
Query: 354 GLY-LIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ + NG+L L+G LDWP R IA+G ARGL YLH +IHRD
Sbjct: 2364 DQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRD 2543
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IK+SN+LL ++ DFG +K+ P + + + V GT GYL PE + + EK+D+
Sbjct: 2544 IKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDV 2723
Query: 473 FAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYR 528
F+FGV+LLEIV+GR P++ ++ +++ WA P + + E+ DP ++G Y E ++R
Sbjct: 2724 FSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWR 2903
Query: 529 VVLTASYCVRQTAIWRPAMTEV 550
VV A C+ + +RP+M +
Sbjct: 2904 VVEVALQCLEPFSTYRPSMVAI 2969
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 186 bits (473), Expect = 6e-48
Identities = 97/263 (36%), Positives = 158/263 (59%), Gaps = 6/263 (2%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ +VYKG L GE +AVK+L+ D + +E F+ME+ ++ + H N L+GYC +
Sbjct: 478 GGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQE--FVMEVLMLSLLHHTNLVRLIGYCTDG 651
Query: 354 GL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L++ Y G+L L L+W R K+A+G ARGL YLH +I+RD
Sbjct: 652 DQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRD 831
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
+K++N+LL ++ P+++DFGLAK P H V GT+GY APE M G + K+DI
Sbjct: 832 LKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDI 1011
Query: 473 FAFGVLLLEIVTGRRPVDSSK----QNILLWAKP-LMESGNIAELADPRMEGRYDVEQLY 527
++FGV+LLE++TGRR +D+S+ QN++ WA+P + + DP ++GR+ L+
Sbjct: 1012YSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLH 1191
Query: 528 RVVLTASYCVRQTAIWRPAMTEV 550
+ + + C+++ +RP +T++
Sbjct: 1192QAIAITAMCLQEQPKFRPLITDI 1260
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 182 bits (461), Expect = 2e-46
Identities = 99/267 (37%), Positives = 156/267 (58%), Gaps = 10/267 (3%)
Frame = +2
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
G Y VY L +G +AVK+L + +P EFL ++ ++ + + N L GYC E
Sbjct: 386 GSYGRVYYATLNDGNAVAVKKLDVSS-EPETNNEFLTQVSMVSRLKNDNFVELHGYCVEG 562
Query: 354 GL-YLIFNYSQNGNLSTALHGY-----GKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHR 407
L L + ++ G+L LHG + G +LDW R +IA+ ARGL YLH+ +
Sbjct: 563 NLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAVDAARGLEYLHEKVQPA 742
Query: 408 IIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVD 467
IIHRDI++SNVL+ DY+ +I DF L+ P+ V GTFGY APE M G +
Sbjct: 743 IIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLT 922
Query: 468 EKTDIFAFGVLLLEIVTGRRPVDSS----KQNILLWAKPLMESGNIAELADPRMEGRYDV 523
+K+D+++FGV+LLE++TGR+PVD + +Q+++ WA P + + + DP+++G Y
Sbjct: 923 QKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPP 1102
Query: 524 EQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ + ++ A+ CV+ A +RP M+ V
Sbjct: 1103KGVAKLAAVAALCVQYEAEFRPNMSIV 1183
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 178 bits (451), Expect = 2e-45
Identities = 106/234 (45%), Positives = 140/234 (59%), Gaps = 6/234 (2%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAK-DNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
GG+ VY G L +G IAVKRL NK + EF +E+ I+ V H N SL GYC E
Sbjct: 91 GGFGSVYWGQLWDGSQIAVKRLKVWSNK---ADMEFAVEVEILARVRHKNLLSLRGYCAE 261
Query: 353 NGLYLI-FNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
LI ++Y N +L + LHG + LDW R IAIG A G+ YLH IIHR
Sbjct: 262 GQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAEGIVYLHHQATPHIIHR 441
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
DIKASNVLL D++ ++ DFG AK +P+ T H V+GT GYLAPE M G +E D
Sbjct: 442 DIKASNVLLDSDFQARVADFGFAKLIPDGAT-HVTTRVKGTLGYLAPEYAMLGKANECCD 618
Query: 472 IFAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLMESGNIAELADPRMEGRY 521
+F+FG+LLLE+ +G++P++ + K++I WA PL + E ADPR+ G Y
Sbjct: 619 VFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKKFTEFADPRLNGEY 780
>CN825263
Length = 663
Score = 173 bits (439), Expect = 6e-44
Identities = 93/217 (42%), Positives = 136/217 (61%), Gaps = 5/217 (2%)
Frame = +1
Query: 295 GYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFENG 354
G+ VYKG L +G +AVK L +D D +EFL E+ ++ + H N L+G C E
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRD--DQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQ 174
Query: 355 LY-LIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
LI+ NG++ + LHG K LDW R KIA+G ARGL YLH+ +IHRD
Sbjct: 175 TRCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDF 354
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
K+SN+LL D+ P+++DFGLA+ ++ H V GTFGYLAPE M G + K+D++
Sbjct: 355 KSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVY 534
Query: 474 AFGVLLLEIVTGRRPVDSS----KQNILLWAKPLMES 506
++GV+LLE++TG +PVD S ++N++ WA+P++ S
Sbjct: 535 SYGVVLLELLTGTKPVDLSQPPGQENLVTWARPILTS 645
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 172 bits (436), Expect = 1e-43
Identities = 99/268 (36%), Positives = 148/268 (54%), Gaps = 10/268 (3%)
Frame = +2
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG VY+G + NG +A+KRL N + F E+ +G + H N LLGY
Sbjct: 2216 KGGAGIVYRGSMPNGTDVAIKRLVGQGSGRN-DYGFRAEIETLGKIRHRNIMRLLGYVSN 2392
Query: 353 NGL-YLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHR 411
L++ Y NG+L LHG G L W +RYKIA+ ARGL Y+H C IIHR
Sbjct: 2393 KDTNLLLYEYMPNGSLGEWLHG--AKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHR 2566
Query: 412 DIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTD 471
D+K++N+LL D+E + DFGLAK+L + ++ + G++GY+APE VDEK+D
Sbjct: 2567 DVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSD 2746
Query: 472 IFAFGVLLLEIVTGRRPVD--SSKQNILLWAKPLM-------ESGNIAELADPRMEGRYD 522
+++FGV+LLE++ GR+PV +I+ W M ++ + + DPR+ G Y
Sbjct: 2747 VYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSG-YP 2923
Query: 523 VEQLYRVVLTASYCVRQTAIWRPAMTEV 550
+ + + A CV++ RP M EV
Sbjct: 2924 LTSVIHMFNIAMMCVKEMGPARPTMREV 3007
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 172 bits (436), Expect = 1e-43
Identities = 97/216 (44%), Positives = 130/216 (59%), Gaps = 5/216 (2%)
Frame = +2
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF-E 352
GG+ VY G +G IAVK+L N E EF +E+ ++G V H N L GYC +
Sbjct: 329 GGFGSVYWGRTSDGLQIAVKKLKAMNS--KAEMEFAVEVEVLGRVRHKNLLGLRGYCVGD 502
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
+ ++++Y N +L + LHG L+W R KIAIG A G+ YLH IIHRD
Sbjct: 503 DQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRD 682
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDI 472
IKASNVLL D+EP + DFG AK +P + H V+GT GYLAPE M G V E D+
Sbjct: 683 IKASNVLLNSDFEPLVADFGFAKLIP-EGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDV 859
Query: 473 FAFGVLLLEIVTGRRPVD----SSKQNILLWAKPLM 504
++FG+LLLE+VTGR+P++ K+ I WA+PL+
Sbjct: 860 YSFGILLLELVTGRKPIEKLPGGVKRTITEWAEPLI 967
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 159 bits (401), Expect = 1e-39
Identities = 95/265 (35%), Positives = 147/265 (54%), Gaps = 8/265 (3%)
Frame = +3
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEK---EFLMELGIIGHVCHPNTASLLGYC 350
GG+ +VY G++ G +A+KR +P E+ EF E+ ++ + H + SL+GYC
Sbjct: 24 GGFGKVYYGEVDGGTKVAIKR-----GNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYC 188
Query: 351 FENG-LYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRII 409
EN + L++++ G L H Y L W R +I IG ARGLHYLH K+ II
Sbjct: 189 EENTEMILVYDHMAYGTLRE--HLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTII 362
Query: 410 HRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEK 469
HRD+K +N+LL + +++DFGL+K P H V+G+FGYL PE F + +K
Sbjct: 363 HRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDK 542
Query: 470 TDIFAFGVLLLEIVTGRRPVDSS--KQNILL--WAKPLMESGNIAELADPRMEGRYDVEQ 525
+D+++FGV+L EI+ R ++ S K+ + L WA G + ++ DP ++G+ E
Sbjct: 543 SDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIAPEC 722
Query: 526 LYRVVLTASYCVRQTAIWRPAMTEV 550
+ TA CV I RP+M +V
Sbjct: 723 FKKFAETAMKCVSDQGIERPSMGDV 797
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 157 bits (396), Expect = 5e-39
Identities = 90/267 (33%), Positives = 143/267 (52%), Gaps = 12/267 (4%)
Frame = +1
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE 352
+GG+ VY +L G+ A+K++ D EFL EL ++ HV H N L+GYC E
Sbjct: 1111 QGGFGAVYYAEL-RGKKTAIKKM-----DVQASTEFLCELKVLTHVHHLNLVRLIGYCVE 1272
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
L+L++ + NGNL LHG GK L W R +IA+ ARGL Y+H+ IHRD
Sbjct: 1273 GSLFLVYEHIDNGNLGQYLHGSGK--EPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRD 1446
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLP--NKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKT 470
+K++N+L+ + ++ DFGL K + N ++ GTFGY+ PE +G + K
Sbjct: 1447 VKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLV---GTFGYMPPEYAQYGDISPKI 1617
Query: 471 DIFAFGVLLLEIVTGRRP-------VDSSKQNILLWAKPLMES---GNIAELADPRMEGR 520
D++AFGV+L E+++ + V SK + L+ + L +S + +L DPR+
Sbjct: 1618 DVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDALRKLVDPRLGEN 1797
Query: 521 YDVEQLYRVVLTASYCVRQTAIWRPAM 547
Y ++ + ++ C R + RP+M
Sbjct: 1798 YPIDSVLKIAQLGRACTRDNPLLRPSM 1878
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 135 bits (340), Expect = 2e-32
Identities = 90/261 (34%), Positives = 132/261 (50%), Gaps = 4/261 (1%)
Frame = +1
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFEN 353
GG+ VYKG L NG+ IAVKRL+ N +EF E+ +I + H N L G
Sbjct: 1699 GGFGPVYKGLLANGQEIAVKRLS--NTSGQGMEEFKNEIKLIARLQHRNLVKLFGCSVHQ 1872
Query: 354 GLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDI 413
+ + + N + +DW R +I G+ARGL YLH+ + RIIHRD+
Sbjct: 1873 ------DENSHANKKMKILLDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDL 2034
Query: 414 KASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIF 473
K SN+LL + P+I+DFGLA+ V GT+GY+ PE +HG K+D+F
Sbjct: 2035 KTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVF 2214
Query: 474 AFGVLLLEIVTGR---RPVDSSKQ-NILLWAKPLMESGNIAELADPRMEGRYDVEQLYRV 529
+FGV++LEI++G+ R D N+L A L EL D ++ ++ R
Sbjct: 2215 SFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEERPLELVDELLDDPVIPTEILRY 2394
Query: 530 VLTASYCVRQTAIWRPAMTEV 550
+ A CV++ RP M +
Sbjct: 2395 IHVALLCVQRRPENRPDMLSI 2457
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 133 bits (335), Expect = 6e-32
Identities = 70/174 (40%), Positives = 104/174 (59%), Gaps = 4/174 (2%)
Frame = +3
Query: 372 HGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDIKASNVLLGPDYEPQITDF 431
H + G L W R KIA+G ARGL YLH+ + IIHR IK+SN+LL D +I DF
Sbjct: 9 HEGAQPGLVLSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIADF 188
Query: 432 GLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDS 491
L+ P+ V GTFGY APE M G + K+D+++FGV+LLE++TGR+PVD
Sbjct: 189 DLSNQAPDAAARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVDH 368
Query: 492 S----KQNILLWAKPLMESGNIAELADPRMEGRYDVEQLYRVVLTASYCVRQTA 541
+ +Q+++ WA P + + + D R++G Y V+ + ++ A+ CV+ A
Sbjct: 369 TLPRGQQSLVTWATPKLSEDKVKQCVDARLKGEYPVKSVAKMAAVAALCVQYEA 530
>BP032158
Length = 555
Score = 133 bits (334), Expect = 8e-32
Identities = 72/162 (44%), Positives = 103/162 (63%), Gaps = 1/162 (0%)
Frame = +2
Query: 294 GGYSEVYKGDLCNGETIAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCFE- 352
GG+ VYKG L G+ IAVK+L+ +K N+E F+ E+G+I + HPN L G C E
Sbjct: 77 GGFGPVYKGVLSEGDVIAVKQLSSKSKQGNRE--FINEIGMISALQHPNLVKLYGCCIEG 250
Query: 353 NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRD 412
N L L++ Y +N +L+ AL G + +L+W R KI +G+A+GL YLH+ + +I+HRD
Sbjct: 251 NQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVGIAKGLAYLHEESRLKIVHRD 430
Query: 413 IKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFG 454
IKA+NVLL D + +I+DFGLAK L + H + GT G
Sbjct: 431 IKATNVLLDKDLKAKISDFGLAK-LDEEENTHISTRIAGTIG 553
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 133 bits (334), Expect = 8e-32
Identities = 86/226 (38%), Positives = 120/226 (53%), Gaps = 6/226 (2%)
Frame = +3
Query: 331 ELGIIGHVCHPNTASLLGYCFE-NGLYLIFNYSQNGNLSTALHGYGKAGNSLDWPIRYKI 389
E+ ++ + H N LLG+ + N LI Y NG L L G G LD+ R +I
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGL--RGKILDFNQRLEI 179
Query: 390 AIGVARGLHYLHKCCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLP-NKWTHHAVIP 448
AI VA GL YLH + +IIHRD+K+SN+LL ++ DFG A+ P N H
Sbjct: 180 AIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK 359
Query: 449 VEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDSSK----QNILLWAKPLM 504
V+GT GYL PE + K+D+++FG+LLLEI+TGRRPV+ K + L WA
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 505 ESGNIAELADPRMEGRYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
G++ EL DP ME + + L +++ + C RP M V
Sbjct: 540 NEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSV 677
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 126 bits (316), Expect = 1e-29
Identities = 92/266 (34%), Positives = 141/266 (52%), Gaps = 8/266 (3%)
Frame = +3
Query: 293 RGGYSEVYKGDLCNGETIAVKRLAKDNKDPN-KEKEFLMELGIIGHVCHPNTASLLGYCF 351
+G + YK L G +AVKRL KD EKEF ++ +G + H + L Y F
Sbjct: 300 KGTFGTAYKAVLETGLVVAVKRL----KDVTISEKEFKDKIETVGAMDHQSLVPLRAYYF 467
Query: 352 ENG-LYLIFNYSQNGNLSTALHGYGKAGNS-LDWPIRYKIAIGVARGLHYLHKCCKHRII 409
L+++Y G+LS LHG AG + L+W IR IA+G ARG+ YLH + +
Sbjct: 468 SRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQGPN-VS 644
Query: 410 HRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEK 469
H +IKASN+LL YE +++DFGLA + T + V GY APEV V +K
Sbjct: 645 HGNIKASNILLTKSYEAKVSDFGLAHLVGPSSTPNRVA------GYRAPEVTDPRKVSQK 806
Query: 470 TDIFAFGVLLLEIVTGRRP----VDSSKQNILLWAKPLMESGNIAELADPRMEGRYDV-E 524
D+++FGVLLLE++TG+ P ++ ++ W + L+ +E+ D + +V E
Sbjct: 807 ADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVREEWTSEVFDLELLRYQNVEE 986
Query: 525 QLYRVVLTASYCVRQTAIWRPAMTEV 550
++ +++ A C RP++ EV
Sbjct: 987 EMVQLLQLAVDCAAPYPDRRPSIAEV 1064
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 123 bits (309), Expect = 7e-29
Identities = 74/211 (35%), Positives = 112/211 (53%), Gaps = 12/211 (5%)
Frame = +2
Query: 352 ENGLYLIFNYSQNGNLSTALHGYGKAGNS---------LDWPIRYKIAIGVARGLHYLHK 402
E + L++ Y +N +L LH K+ + LDWP R KIAIG A+GL Y+H
Sbjct: 38 EASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAIGAAQGLSYMHH 217
Query: 403 CCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNKWTHHAVIPVEGTFGYLAPEVFM 462
C I+HRD+K SN+LL + ++ DFGLA+ L + + V GTFGY+APE
Sbjct: 218 DCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIGTFGYIAPEYVQ 397
Query: 463 HGIVDEKTDIFAFGVLLLEIVTGRRPVDSSKQNILL-WA-KPLMESGNIAE-LADPRMEG 519
+ EK D+++FGV+LLE+ TG+ + + L WA + ++ N+ + L ME
Sbjct: 398 TTRISEKVDVYSFGVVLLELTTGKEANYGDQHSSLAEWAWRHILIGSNVXDLLXKDVMEA 577
Query: 520 RYDVEQLYRVVLTASYCVRQTAIWRPAMTEV 550
Y ++++ V C RP+M EV
Sbjct: 578 SY-IDEMCSVFKLGVMCTATLPATRPSMKEV 667
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 120 bits (301), Expect = 6e-28
Identities = 70/177 (39%), Positives = 104/177 (58%), Gaps = 5/177 (2%)
Frame = +3
Query: 293 RGGYSEVYKGDLCNGET-IAVKRLAKDNKDPNKEKEFLMELGIIGHVCHPNTASLLGYCF 351
+GGY VY+G L + +AVK ++D +FL EL II + H + L G+C
Sbjct: 192 QGGYGVVYRGMLPKEKLEVAVKMFSRDKM--KSTDDFLSELIIINRLRHKHLVRLQGWCH 365
Query: 352 ENG-LYLIFNYSQNGNLSTAL--HGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRI 408
+NG L L+++Y NG+L + + G L WP+RYKI GVA L+YLH ++
Sbjct: 366 KNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKIISGVASALNYLHNEYDQKV 545
Query: 409 IHRDIKASNVLLGPDYEPQITDFGLAKWLPN-KWTHHAVIPVEGTFGYLAPEVFMHG 464
+HRD+KASN++L ++ ++ DFGLA+ L N K ++ + V GT GY+APE F G
Sbjct: 546 VHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEGVHGTMGYIAPECFHTG 716
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 119 bits (298), Expect = 1e-27
Identities = 72/202 (35%), Positives = 110/202 (53%), Gaps = 17/202 (8%)
Frame = +3
Query: 331 ELGIIGHVCHPNTASLLGYCF--ENGLYLIFNYSQNGNLSTALHGYGKA--------GNS 380
E+ I+ ++ H N L C E L L+++Y +N +L LH K N
Sbjct: 3 EVEILSNIRHNNIVKLQ-CCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNI 179
Query: 381 LDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDIKASNVLLGPDYEPQITDFGLAKWLPNK 440
+DWP R IAIGVA+GL Y+H C ++HRD+K SN+LL + ++ DFGLA
Sbjct: 180 IDWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKP 359
Query: 441 WTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDSSKQNILL-W 499
+ V GTFGY+APE + V+EK D+++FGV+LLE+ TG+ + + L W
Sbjct: 360 EELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGKEANHGDEYSSLAEW 539
Query: 500 AKPLMESGN------IAELADP 515
A+ ++ G+ + E+A+P
Sbjct: 540 ARRHIQIGSNIEDLLVEEIAEP 605
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 116 bits (291), Expect = 8e-27
Identities = 62/152 (40%), Positives = 94/152 (61%), Gaps = 5/152 (3%)
Frame = +3
Query: 372 HGYGKAGNSLDWPIRYKIAIGVARGLHYLHKCCKHRIIHRDIKASNVLLGPDYEPQITDF 431
H + + L W +R K+AIG ARGL +LH K ++I+RD KASN+LL ++ +++DF
Sbjct: 30 HLFRRGPQPLSWSVRMKVAIGAARGLSFLHNA-KSQVIYRDFKASNILLDAEFNAKLSDF 206
Query: 432 GLAKWLPNKWTHHAVIPVEGTFGYLAPEVFMHGIVDEKTDIFAFGVLLLEIVTGRRPVDS 491
GLAK P H V GT GY APE G + K+D+++FGV+LLE+++GRR VD
Sbjct: 207 GLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDK 386
Query: 492 S----KQNILLWAKP-LMESGNIAELADPRME 518
+ QN++ WA+P L + + + D ++E
Sbjct: 387 TIAGVDQNLVDWARPYLGDKRRLFRIMDSKLE 482
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.337 0.149 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,317,604
Number of Sequences: 28460
Number of extensions: 148054
Number of successful extensions: 1579
Number of sequences better than 10.0: 285
Number of HSP's better than 10.0 without gapping: 1414
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1425
length of query: 550
length of database: 4,897,600
effective HSP length: 95
effective length of query: 455
effective length of database: 2,193,900
effective search space: 998224500
effective search space used: 998224500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC127169.5


