

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.12 + phase: 0
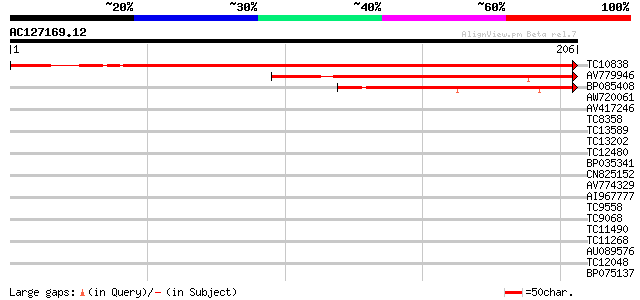
(206 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10838 weakly similar to PIR|T04232|T04232 pathogenesis-related... 335 3e-93
AV779946 153 2e-38
BP085408 88 1e-18
AW720061 35 0.007
AV417246 30 0.30
TC8358 similar to UP|Q84T14 (Q84T14) Vacuolar ATPase subunit E (... 29 0.66
TC13589 28 0.87
TC13202 28 0.87
TC12480 28 1.1
BP035341 28 1.1
CN825152 28 1.5
AV774329 28 1.5
AI967777 28 1.5
TC9558 similar to GB|BAC65066.1|28812162|AP005847 plastid RNA po... 27 1.9
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 27 1.9
TC11490 similar to UP|O48566 (O48566) CLP protease regulatory su... 27 1.9
TC11268 similar to UP|Q7SBH0 (Q7SBH0) Predicted protein, partial... 27 1.9
AU089576 27 2.5
TC12048 weakly similar to UP|Q9LGA3 (Q9LGA3) ESTs D22655(C0749),... 27 3.3
BP075137 27 3.3
>TC10838 weakly similar to PIR|T04232|T04232 pathogenesis-related protein
homolog F14M19.60 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (78%)
Length = 914
Score = 335 bits (859), Expect = 3e-93
Identities = 152/206 (73%), Positives = 173/206 (83%)
Frame = +3
Query: 1 MKSHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQL 60
M++HL+L F + + P S+SS P Q +NQ L+ +K PDNE+IYKVSKQL
Sbjct: 201 MRAHLILFLFLLVLA----------NPQSASSVP-QTHNQ-LTPKKIPDNETIYKVSKQL 344
Query: 61 CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK 120
CW C+ ESLEFLFRHN+VRA+KWE PLMWD+QL+ YARWWA QRKPDCKVEHSFPE+ FK
Sbjct: 345 CWGCIGESLEFLFRHNMVRAAKWESPLMWDFQLQSYARWWAGQRKPDCKVEHSFPENDFK 524
Query: 121 LGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR 180
LGENI+WGSGS WTPTDAVKAWADEEKYYTY TN+C GQMCGHYTQIVWK+T+R+GCAR
Sbjct: 525 LGENIFWGSGSAWTPTDAVKAWADEEKYYTYATNTCEEGQMCGHYTQIVWKNTKRVGCAR 704
Query: 181 VVCDDGDVFMTCNYDPVGNYVGERPY 206
VVCDDGDVFMTCNYDPVGNYVGERPY
Sbjct: 705 VVCDDGDVFMTCNYDPVGNYVGERPY 782
>AV779946
Length = 533
Score = 153 bits (387), Expect = 2e-38
Identities = 66/112 (58%), Positives = 81/112 (71%), Gaps = 1/112 (0%)
Frame = -3
Query: 96 YARWWASQRKPDCKVEHSFPEDGFKLGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNS 155
YA+W+A+QR+ DC +EHS GENI+WGSG+ W P+ AV AW +E ++Y Y NS
Sbjct: 528 YAQWYANQRRNDCALEHS----NGPYGENIFWGSGTGWKPSPAVDAWVEERQWYNYWHNS 361
Query: 156 CVSGQMCGHYTQIVWKSTRRIGCARVVCDDGD-VFMTCNYDPVGNYVGERPY 206
C +G+MCGHYTQIVW TR++GCA V C G FMTCNYDP GNY GERPY
Sbjct: 360 CANGEMCGHYTQIVWGDTRKVGCASVTCSGGQGTFMTCNYDPPGNYYGERPY 205
>BP085408
Length = 350
Score = 87.8 bits (216), Expect = 1e-18
Identities = 41/89 (46%), Positives = 59/89 (66%), Gaps = 2/89 (2%)
Frame = -1
Query: 120 KLGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQM-CGHYTQIVWKSTRRIGC 178
K G N G G WTP+ AV+ W +E+++Y+Y TNSCV+ C HYTQ+VW+ ++++GC
Sbjct: 347 KYGSNQDLG-GMGWTPSVAVQDWVNEKEFYSYRTNSCVAPSFPCWHYTQVVWRKSKQVGC 171
Query: 179 ARVVCDDGDVFMT-CNYDPVGNYVGERPY 206
A++ C + +T C YDP GN GE PY
Sbjct: 170 AQLTCVVDKLILTICFYDPPGNINGESPY 84
>AW720061
Length = 304
Score = 35.4 bits (80), Expect = 0.007
Identities = 11/25 (44%), Positives = 18/25 (72%)
Frame = +3
Query: 159 GQMCGHYTQIVWKSTRRIGCARVVC 183
G CG YTQ+VW+ ++ +GC++ C
Sbjct: 207 GT*CGVYTQVVWRDSKEVGCSQASC 281
Score = 33.9 bits (76), Expect = 0.021
Identities = 20/59 (33%), Positives = 27/59 (44%)
Frame = +1
Query: 122 GENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR 180
G I GS S P AV W E+ +Y + NSCV+ G ++ RR+G R
Sbjct: 100 GNQIMAGSVSV-APRVAVAEWVKEKDFYIHANNSCVAEHSVGFTRRLCGGIRRRLGARR 273
>AV417246
Length = 370
Score = 30.0 bits (66), Expect = 0.30
Identities = 21/72 (29%), Positives = 38/72 (52%)
Frame = +1
Query: 13 FVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWNCMQESLEFL 72
F+ F + + S + + +S SPT +NQ LSQQ++ +++ ++ L +E LE L
Sbjct: 13 FIAFPRQNICQSAR-TGTSHSPTNHWNQSLSQQQRMCLKAVPQLI--LTPQSQREPLEKL 183
Query: 73 FRHNLVRASKWE 84
+ LV W+
Sbjct: 184MLNQLVARMDWQ 219
>TC8358 similar to UP|Q84T14 (Q84T14) Vacuolar ATPase subunit E (Fragment),
complete
Length = 1321
Score = 28.9 bits (63), Expect = 0.66
Identities = 17/75 (22%), Positives = 35/75 (46%)
Frame = -3
Query: 7 LLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWNCMQ 66
L + + F + + + S +SSS+ T+I + S ++++ +CW C++
Sbjct: 260 LRSYSCLIFFFSASTNCSFSMLNSSSAETEISLAFSSAS--------WRMNLTICWICLE 105
Query: 67 ESLEFLFRHNLVRAS 81
S F+F H +S
Sbjct: 104 TSPSFIFAHRKTTSS 60
>TC13589
Length = 447
Score = 28.5 bits (62), Expect = 0.87
Identities = 14/42 (33%), Positives = 25/42 (59%)
Frame = -3
Query: 18 TKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQ 59
T +LS+SP PS+ +SSP+ + ++ P + S K+ +Q
Sbjct: 376 TPSLSSSPSPSAQTSSPSGSH*PPMTSASAPASTSARKLCQQ 251
>TC13202
Length = 431
Score = 28.5 bits (62), Expect = 0.87
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = +1
Query: 3 SHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQ 36
SH LLL F F++ TL+ +P PS ++ TQ
Sbjct: 16 SHFLLLLHFSAPPFSSATLNHTPPPSPRMAAATQ 117
>TC12480
Length = 785
Score = 28.1 bits (61), Expect = 1.1
Identities = 14/64 (21%), Positives = 28/64 (42%), Gaps = 11/64 (17%)
Frame = +2
Query: 17 TTKTLSTSPQPSSSSSSPTQIYN-----------QYLSQQKKPDNESIYKVSKQLCWNCM 65
+T + P + SS+PT+ N +++ K+P+ ++ +L WN
Sbjct: 365 STSVTNLPADPHAGSSNPTEFVNHGLTLWNQTRQRWIGNNKRPEKQTEQSREPKLSWNAT 544
Query: 66 QESL 69
+SL
Sbjct: 545 YDSL 556
>BP035341
Length = 523
Score = 28.1 bits (61), Expect = 1.1
Identities = 14/36 (38%), Positives = 20/36 (54%), Gaps = 2/36 (5%)
Frame = -2
Query: 161 MCGHYTQIVWKSTRRIGCA--RVVCDDGDVFMTCNY 194
+C H V KS + A ++VCD GD F +C+Y
Sbjct: 183 VCVHVLVFVLKSMDNVFLAYSKLVCDIGDNFFSCDY 76
>CN825152
Length = 697
Score = 27.7 bits (60), Expect = 1.5
Identities = 27/106 (25%), Positives = 37/106 (34%), Gaps = 8/106 (7%)
Frame = -1
Query: 74 RHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVE-HSFPEDGFKLGENIYWGSGSD 132
RH+L R L L + A +W D + HS G + E G G
Sbjct: 316 RHDLSRRRSLFLSLFYRIGFHGGAAFWILHEDTDGEARSHSSGARG*AVMETSQGGGGEG 137
Query: 133 ----WTPTDAVKAW---ADEEKYYTYVTNSCVSGQMCGHYTQIVWK 171
W + + W A EE + Y G+ C H +VWK
Sbjct: 136 ECWRWVEEEGERRWLMVAAEEGLHGYG-----GGESCRHSRSLVWK 14
>AV774329
Length = 443
Score = 27.7 bits (60), Expect = 1.5
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = +3
Query: 3 SHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDN 50
S++L++ FIF F S+S S+SSS+P NQ + P+N
Sbjct: 225 SNVLIIFIFIFFPFIL-LQSSSSSSSNSSSNPENEKNQEWPNIENPEN 365
>AI967777
Length = 461
Score = 27.7 bits (60), Expect = 1.5
Identities = 17/53 (32%), Positives = 22/53 (41%), Gaps = 6/53 (11%)
Frame = +1
Query: 149 YTYVTNSCVSGQMCGHYTQIVWKSTRRIGC------ARVVCDDGDVFMTCNYD 195
Y TNSC+ G C ++ I S GC + C DG C+YD
Sbjct: 100 YGSKTNSCIEGHPCWRHSSIC*SSRCNEGCRWCKNWSSCSCHDGG--SDCSYD 252
>TC9558 similar to GB|BAC65066.1|28812162|AP005847 plastid RNA polymerase
sigma factor {Oryza sativa (japonica cultivar-group);} ,
partial (4%)
Length = 696
Score = 27.3 bits (59), Expect = 1.9
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = -3
Query: 22 STSPQPSSSSSSPTQIYNQYLSQQKKP 48
S +P PSSS S+PT Y+ + SQ P
Sbjct: 247 SPTPSPSSS*SNPTTHYSFFCSQTTAP 167
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 27.3 bits (59), Expect = 1.9
Identities = 17/54 (31%), Positives = 28/54 (51%)
Frame = -1
Query: 11 FIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWNC 64
F+ + ++ + S+S SSSSSS + L+++ KPD + VS C C
Sbjct: 165 FLSPSSSSSSSSSSSSSSSSSSSSSSSSGLGLNKRLKPDGAVLILVSCCCCCCC 4
>TC11490 similar to UP|O48566 (O48566) CLP protease regulatory subunit CLPX
precursor, partial (4%)
Length = 528
Score = 27.3 bits (59), Expect = 1.9
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = +2
Query: 11 FIFVTFTTKTLSTSPQPSSSS 31
F+ T TT T+STSP PS+++
Sbjct: 215 FLHATATTTTISTSPPPSTAA 277
Score = 26.6 bits (57), Expect = 3.3
Identities = 8/25 (32%), Positives = 18/25 (72%)
Frame = -2
Query: 81 SKWELPLMWDYQLEQYARWWASQRK 105
+KW+L L+W ++ E+ +WW ++ +
Sbjct: 257 AKWKLLLLWRWR-EERVKWWVAREE 186
>TC11268 similar to UP|Q7SBH0 (Q7SBH0) Predicted protein, partial (5%)
Length = 1097
Score = 27.3 bits (59), Expect = 1.9
Identities = 14/32 (43%), Positives = 21/32 (64%)
Frame = -2
Query: 24 SPQPSSSSSSPTQIYNQYLSQQKKPDNESIYK 55
+P+PSSSSS+ I+N L + +K N+S K
Sbjct: 898 NPEPSSSSSNTFIIFNIKLKKIQKTKNQSPLK 803
>AU089576
Length = 353
Score = 26.9 bits (58), Expect = 2.5
Identities = 8/28 (28%), Positives = 17/28 (60%)
Frame = -1
Query: 54 YKVSKQLCWNCMQESLEFLFRHNLVRAS 81
++++ +CW C++ S F+F H +S
Sbjct: 107 WRMNLTICWICLETSPSFIFAHRKTTSS 24
>TC12048 weakly similar to UP|Q9LGA3 (Q9LGA3) ESTs D22655(C0749), partial
(20%)
Length = 697
Score = 26.6 bits (57), Expect = 3.3
Identities = 16/31 (51%), Positives = 19/31 (60%)
Frame = -2
Query: 3 SHLLLLCFFIFVTFTTKTLSTSPQPSSSSSS 33
S + L FF F TFT T ST P ++SSSS
Sbjct: 456 SATVTLGFFGFFTFTFFTSSTFPFSATSSSS 364
>BP075137
Length = 507
Score = 26.6 bits (57), Expect = 3.3
Identities = 11/29 (37%), Positives = 16/29 (54%)
Frame = -3
Query: 8 LCFFIFVTFTTKTLSTSPQPSSSSSSPTQ 36
LCFF+F++ T T S + S + TQ
Sbjct: 109 LCFFVFISLTRSTFSFLRSSNEMSEAMTQ 23
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.133 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,226,701
Number of Sequences: 28460
Number of extensions: 94303
Number of successful extensions: 838
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 802
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 824
length of query: 206
length of database: 4,897,600
effective HSP length: 86
effective length of query: 120
effective length of database: 2,450,040
effective search space: 294004800
effective search space used: 294004800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC127169.12


