

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127021.9 + phase: 0
(540 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
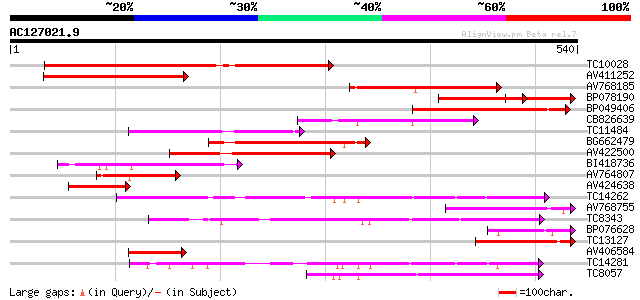
Score E
Sequences producing significant alignments: (bits) Value
TC10028 weakly similar to UP|Q9LXX7 (Q9LXX7) Cytochrome P450-lik... 415 e-117
AV411252 233 6e-62
AV768185 225 1e-59
BP078190 176 1e-44
BP049406 138 2e-33
CB826639 132 1e-31
TC11484 similar to PIR|JC5965|JC5965 cytochrome P450 CYP86A1 - A... 132 2e-31
BG662479 119 1e-27
AV422500 117 5e-27
BI418736 105 1e-23
AV764807 99 2e-21
AV424638 94 4e-20
TC14262 UP|Q9MBE5 (Q9MBE5) Cytochrome P450, complete 89 1e-18
AV768755 84 8e-17
TC8343 weakly similar to GB|BAB02401.1|9294391|AB023038 cytochro... 81 4e-16
BP076628 76 2e-14
TC13127 weakly similar to UP|O49373 (O49373) Cytochrome P450 - l... 74 5e-14
AV406584 74 6e-14
TC14281 similar to UP|Q9XHC6 (Q9XHC6) Cytochrome P450 monooxygen... 73 1e-13
TC8057 weakly similar to UP|Q8W228 (Q8W228) Cytochrome P450, par... 72 2e-13
>TC10028 weakly similar to UP|Q9LXX7 (Q9LXX7) Cytochrome P450-like protein,
partial (16%)
Length = 874
Score = 415 bits (1067), Expect = e-117
Identities = 204/275 (74%), Positives = 235/275 (85%)
Frame = +1
Query: 34 SLQSTMSTTFTFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVDY 93
SLQS + T +FLFFSFTLLFS FSFL+FISR+KPWCNC+ CK++LTMSW++KF N D+
Sbjct: 79 SLQSMSAATCSFLFFSFTLLFSLFSFLIFISRMKPWCNCDLCKSYLTMSWTSKFFNFCDW 258
Query: 94 YTHLLQESPTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFN 153
YTHLL+ SPTGTIHVHVL NTITSNPENVE+ILKTNF NYPKGK FSTILGDLLGRGIFN
Sbjct: 259 YTHLLRTSPTGTIHVHVLKNTITSNPENVEHILKTNFQNYPKGKPFSTILGDLLGRGIFN 438
Query: 154 VDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSED 213
VDG SWKFQRKMAS+ELGSV IRSYAMELV EEI+TRL PL+A+ TA+ A+N
Sbjct: 439 VDGESWKFQRKMASMELGSVAIRSYAMELVNEEIQTRLTPLMAA-----TAASANN---- 591
Query: 214 VLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLI 273
+LD+QDILRRFSFDNICKFSFGLDPCCL+PS PVS LA AFDL+STLSAQRA+ AS +
Sbjct: 592 -VLDLQDILRRFSFDNICKFSFGLDPCCLLPSFPVSELAEAFDLASTLSAQRAMAASSFV 768
Query: 274 WKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRRE 308
WK KRFFN+G+EKKLKEA+ +VN A +MI ++RE
Sbjct: 769 WKAKRFFNLGNEKKLKEAVGVVNHFAQKMINEKRE 873
>AV411252
Length = 429
Score = 233 bits (594), Expect = 6e-62
Identities = 107/138 (77%), Positives = 120/138 (86%)
Frame = +2
Query: 33 LSLQSTMSTTFTFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVD 92
LS +M TT FLFFSFT++FS FSFL+FISRIKPWCNC+TC TF+T SWS+KF NL D
Sbjct: 14 LSCSQSMPTTTAFLFFSFTIVFSAFSFLIFISRIKPWCNCDTCTTFMTASWSSKFTNLCD 193
Query: 93 YYTHLLQESPTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIF 152
+YTHLL++SPTGTIHVHVL NTIT NP NVE++LKTNF+NYPKGK FS ILGDLLGRGIF
Sbjct: 194 WYTHLLRDSPTGTIHVHVLNNTITCNPHNVEHMLKTNFHNYPKGKPFSAILGDLLGRGIF 373
Query: 153 NVDGHSWKFQRKMASLEL 170
NVDGH W FQRKMASLEL
Sbjct: 374 NVDGHEWSFQRKMASLEL 427
>AV768185
Length = 446
Score = 225 bits (574), Expect = 1e-59
Identities = 113/149 (75%), Positives = 126/149 (83%), Gaps = 4/149 (2%)
Frame = +3
Query: 324 MGALNSHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQ 383
MG++N DD+YLRDIVVSFLLAGRDTVAS LT FF LLS +PKVE IR E+DRVM P Q
Sbjct: 3 MGSVND-DDKYLRDIVVSFLLAGRDTVASGLTCFFWLLSNHPKVEALIREEVDRVMIPKQ 179
Query: 384 EC----ATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYH 439
A+FEQ REM YLN AI+ESMRL+PPVQFDSKFA EDDVLPDGTF++KG+RVTYH
Sbjct: 180 NSVQGFASFEQLREMQYLNAAIYESMRLYPPVQFDSKFAHEDDVLPDGTFVRKGTRVTYH 359
Query: 440 PYAMGRMENIWGPDCLEFKPERWLKDGVF 468
PYAMGRME +WG DCLEF+PERWLKDGVF
Sbjct: 360 PYAMGRMERVWGLDCLEFRPERWLKDGVF 446
>BP078190
Length = 445
Score = 176 bits (445), Expect = 1e-44
Identities = 77/85 (90%), Positives = 82/85 (95%)
Frame = -3
Query: 409 PPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWLKDGVF 468
PPVQFDSK+A EDDVLPDGTF++KGSRVTYHPYAMGRME IWGPDCLEFKPERWLKDGVF
Sbjct: 443 PPVQFDSKYATEDDVLPDGTFVRKGSRVTYHPYAMGRMERIWGPDCLEFKPERWLKDGVF 264
Query: 469 VPKCPFKYPVFQAGSRVCLGKELAI 493
VPKCPFKYPVFQAG RVCLGK+LA+
Sbjct: 263 VPKCPFKYPVFQAGVRVCLGKDLAL 189
Score = 81.6 bits (200), Expect = 3e-16
Identities = 40/67 (59%), Positives = 50/67 (73%)
Frame = -1
Query: 473 PFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRGGL 532
P F+ G G+ +VEMK V A+LV+RFD+RVVGP+QE +FAPGLTA+FRGGL
Sbjct: 250 PLSTRFFRLG*GFVWGRIWPLVEMKCVAAALVRRFDIRVVGPDQELRFAPGLTATFRGGL 71
Query: 533 PVKIYER 539
PVK+YER
Sbjct: 70 PVKVYER 50
>BP049406
Length = 566
Score = 138 bits (348), Expect = 2e-33
Identities = 71/153 (46%), Positives = 97/153 (62%), Gaps = 2/153 (1%)
Frame = -1
Query: 384 ECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAM 443
E FE+ + YL A+ E++RL+P V DSK + DDVLPDGTF+ GS VTY Y+
Sbjct: 500 EPLVFEEVDRLVYLKAALSETLRLYPSVPEDSKHVVADDVLPDGTFVPAGSSVTYSIYSA 321
Query: 444 GRMENIWGPDCLEFKPERWLK-DGV-FVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVA 501
GRM + WG DC+EF+PERWL DG FV PFK+ F AG R+CLGK+LA ++MKS+ +
Sbjct: 320 GRMRSTWGEDCMEFRPERWLSLDGTKFVQHDPFKFVAFNAGPRICLGKDLAYLQMKSIAS 141
Query: 502 SLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPV 534
+++ + VV P + + LT + GL V
Sbjct: 140 AVLLNHRLEVV-PGHQVEQKMSLTLFMKNGLKV 45
>CB826639
Length = 542
Score = 132 bits (332), Expect = 1e-31
Identities = 79/185 (42%), Positives = 109/185 (58%), Gaps = 13/185 (7%)
Frame = +3
Query: 275 KMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIENGVESRKDLLSRFMGALNS----H 330
+ ++FF G+EKKLKE++KIV + NE + R DLLSRFM ++
Sbjct: 3 RFQKFFCFGAEKKLKESLKIVENYMNEAVSARE-----ASPSDDLLSRFMRKRDTAGKPF 167
Query: 331 DDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPN-------- 382
D LR I ++F+LAGRDT + AL+ FF L+ +P VEEKI +EL V+
Sbjct: 168 DAAKLRHIALNFVLAGRDTSSVALSWFFWLVMNHPLVEEKIVLELAAVLTDTRGEETGKW 347
Query: 383 -QECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPY 441
+E FE+ + YL A+ E++RL+P V D K+AL DDVLPDGT + GS VTY Y
Sbjct: 348 VEEAVDFEEADRLVYLKAALAETLRLYPSVPEDFKYALNDDVLPDGTVVPAGSTVTYSIY 527
Query: 442 AMGRM 446
++GRM
Sbjct: 528 SVGRM 542
>TC11484 similar to PIR|JC5965|JC5965 cytochrome P450 CYP86A1 - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (43%)
Length = 752
Score = 132 bits (331), Expect = 2e-31
Identities = 71/167 (42%), Positives = 100/167 (59%)
Frame = +1
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+TS+P+N+E+IL+T F+NYPKG ++ T DLLG GIFN DG +W QRK A+LE +
Sbjct: 286 TVTSHPKNIEHILRTRFDNYPKGPRWQTAFNDLLGHGIFNSDGETWLVQRKTAALEFTTR 465
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK RL ++ A++K V +D+QD+L R +FDNIC
Sbjct: 466 TLRQAMARWVNRTIKNRLWCILDKAAKEK-----------VQVDLQDLLLRLTFDNICGL 612
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFF 280
+FG DP L P LP + + AFD + + QR L LIW+ ++FF
Sbjct: 613 TFGKDPETLSPDLPENPFSVAFDTVTETTMQRLLYPG-LIWRFQKFF 750
>BG662479
Length = 462
Score = 119 bits (298), Expect = 1e-27
Identities = 65/158 (41%), Positives = 104/158 (65%), Gaps = 4/158 (2%)
Frame = +1
Query: 190 RLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVS 249
RL+P+++ KT L D QDIL+RF+FDNIC+ +FG DP L+PSLP +
Sbjct: 22 RLIPILSQAELNKTT----------LPDFQDILQRFTFDNICRIAFGFDPEYLLPSLPET 171
Query: 250 NLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRRE- 308
A AFD + +S++R A PL+WK+ N+GSEK+L+EAI V LA ++++++++
Sbjct: 172 AFAKAFDDGTRISSERFNAAVPLLWKLNHLLNVGSEKRLREAIAEVRGLAGKIVREKKQH 351
Query: 309 IENGVESRK---DLLSRFMGALNSHDDEYLRDIVVSFL 343
+++ ES DLLSRF+ + +S D++++ DIV+SF+
Sbjct: 352 LQSKPESEPESLDLLSRFLRSGHS-DEQFVTDIVISFI 462
>AV422500
Length = 446
Score = 117 bits (293), Expect = 5e-27
Identities = 62/159 (38%), Positives = 99/159 (61%), Gaps = 1/159 (0%)
Frame = -3
Query: 153 NVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSE 212
NVDG W QRK+AS +R + + EE++ LLP + +AE E
Sbjct: 444 NVDGEPWLAQRKLASHVFTPRSVRQFVTTTLEEEVRENLLPAMEVLAE-----------E 298
Query: 213 DVLLDMQDILRRFSFDNICKFSFGLDPCC-LVPSLPVSNLANAFDLSSTLSAQRALTASP 271
+ ++D+Q++L RFSF IC+F+ G D CC L PS P+S L AFD+++ +SA+R
Sbjct: 297 ERVVDLQELLGRFSFSVICRFTLGSDVCCCLDPSSPISPLTRAFDVAAEVSARRGAAPLF 118
Query: 272 LIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIE 310
L+WK+KR+ +GSE++LKEA++ V+ L +MI++R+ ++
Sbjct: 117 LVWKVKRWLGVGSERRLKEAVEDVHRLVMKMIQERKVVK 1
>BI418736
Length = 544
Score = 105 bits (263), Expect = 1e-23
Identities = 70/188 (37%), Positives = 95/188 (50%), Gaps = 12/188 (6%)
Frame = +1
Query: 46 LFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWS--NKFVNL-------VDYYTH 96
LFF F LLF FF F P NT L ++ ++ + + Y++
Sbjct: 25 LFFIFPLLFFFF----FFKSSSPSSPTNTNTITLPKAYPLIGSYLAIKANAHCRIQYFSD 192
Query: 97 LLQESPTGTIHVH-VLGNT--ITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFN 153
L+Q SP T H LG T NP V++ILKT+F+ Y KG T L D LG GIFN
Sbjct: 193 LVQISPAATFTFHRPLGRPTIFTGNPATVQHILKTHFSKYQKGSTLVTTLSDFLGSGIFN 372
Query: 154 VDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSED 213
DG +WKFQR++AS E + +R++ +V E+ RL+P++A AE KT
Sbjct: 373 TDGENWKFQRQVASHEFNTKSLRNFVEHVVDAELSNRLVPILAGAAENKTT--------- 525
Query: 214 VLLDMQDI 221
LD QDI
Sbjct: 526 --LDFQDI 543
>AV764807
Length = 529
Score = 99.0 bits (245), Expect = 2e-21
Identities = 49/83 (59%), Positives = 60/83 (72%), Gaps = 3/83 (3%)
Frame = +1
Query: 83 WSNKFVNLVDYYTHLLQESPTGTIHVHVLG---NTITSNPENVEYILKTNFNNYPKGKQF 139
+ N+F L+D+Y LL +SPT TI VH LG +TSNP+NVE+ILKTNF+N+PKGK F
Sbjct: 283 YKNRF-RLLDWYAELLAQSPTNTIVVHRLGARRTVVTSNPQNVEHILKTNFHNFPKGKPF 459
Query: 140 STILGDLLGRGIFNVDGHSWKFQ 162
+ IL D LG GIFNVDG W Q
Sbjct: 460 TDILSDFLGHGIFNVDGEPWLAQ 528
>AV424638
Length = 227
Score = 94.4 bits (233), Expect = 4e-20
Identities = 40/59 (67%), Positives = 48/59 (80%)
Frame = +2
Query: 57 FSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQESPTGTIHVHVLGNTI 115
FSFL+FIS PWCNC+ C ++LTMSW++ F N D+YTHLL+ SPTGTIHVHVL NTI
Sbjct: 50 FSFLIFISGTNPWCNCDLCNSYLTMSWTS*FFNFCDWYTHLLRTSPTGTIHVHVLKNTI 226
>TC14262 UP|Q9MBE5 (Q9MBE5) Cytochrome P450, complete
Length = 1960
Score = 89.4 bits (220), Expect = 1e-18
Identities = 100/425 (23%), Positives = 188/425 (43%), Gaps = 12/425 (2%)
Frame = +1
Query: 102 PTGTIHVHVLGNTITSNPENVEYILKTN-FNNYPKGKQFSTILGDLLGRGIFNVD-GHSW 159
P T++ + + S PE + L+T+ ++ Q S I + V W
Sbjct: 283 PLYTLYFGSMPTVVASTPELFKLFLQTHEATSFSTRFQTSAIRRLTYDNSVAMVPFAPYW 462
Query: 160 KFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQ 219
KF RK+ +L + + L +EI+ ++ ++A+ + K N +E++L
Sbjct: 463 KFIRKVIMNDLLNATTVNKLRPLRSQEIRK----VLKAMAQSAESQKPLNVTEELL---- 618
Query: 220 DILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRF 279
+++ I + G V + + L+ + + L ++
Sbjct: 619 ----KWTNSTISRMMLGEAEHVKDIVREVLKIFGEYSLTDFIWPLKKLRVGQYEKRIDEI 786
Query: 280 FNIGSEKKLKEAIKIVNDLANEMIKQRRE----IENGVESRK--DLLSRFMGALNSH--- 330
FN K I+ V E+IK+R+E +E G +S D L + N
Sbjct: 787 FN-----KFDPVIEKVIKKRQEIIKRRKERNGELEEGEQSVVFLDTLLEYAADENMEIKI 951
Query: 331 DDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQ 390
E ++ +VV F AG D+ A A L NP+V +K R E+D V+ ++
Sbjct: 952 TKEQIKGLVVDFFSAGTDSTAVATDWALAELINNPRVLKKAREEVDSVVGKDR-LVDESD 1128
Query: 391 TREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIW 450
+ + Y+ + E+ R+ PP+ + +++ L +G I +G+ V ++ +A+ R W
Sbjct: 1129IQNLPYIRAIVKETFRMHPPLPVVKRKCVQECEL-NGYVIPEGALVLFNVWAVQRDPKYW 1305
Query: 451 GPDCLEFKPERWLKDGVFVPKCP-FKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDV 509
LEF+PER+L++ K F+ F +G R+C G LA M ++++S+++ F++
Sbjct: 1306KTP-LEFRPERFLEEADIDLKVQHFELLPFGSGRRMCPGVNLATAGMATLLSSVIQCFEL 1482
Query: 510 RVVGP 514
+VVGP
Sbjct: 1483QVVGP 1497
>AV768755
Length = 488
Score = 83.6 bits (205), Expect = 8e-17
Identities = 44/127 (34%), Positives = 71/127 (55%), Gaps = 3/127 (2%)
Frame = -1
Query: 416 KFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWLKD-GVFVPKCPF 474
K+ + LP G + +++ Y Y+MGRME IWG D LEFKPERW+ + G +
Sbjct: 488 KYVINHATLPSGHHVGPNTKILYLLYSMGRMEQIWGDDSLEFKPERWISEKGHSIHMPSS 309
Query: 475 KYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTA--SFRGGL 532
K+ F AG R CLGK + + +K V A+++ +F ++V+ ++ P L+ R G
Sbjct: 308 KFITFNAGPRSCLGKGIPFIMLKMVAAAIIPKFRIQVM---EDHPITPRLSVVMGMRYGF 138
Query: 533 PVKIYER 539
V++ +R
Sbjct: 137 KVQVTKR 117
>TC8343 weakly similar to GB|BAB02401.1|9294391|AB023038 cytochrome P450
{Arabidopsis thaliana;} , partial (60%)
Length = 1784
Score = 81.3 bits (199), Expect = 4e-16
Identities = 88/391 (22%), Positives = 159/391 (40%), Gaps = 14/391 (3%)
Frame = +1
Query: 133 YPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLL 192
Y K ++ L LL G N DG W RK+ S +E++K L
Sbjct: 409 YEFQKPDTSPLFKLLASGFANYDGDKWAKHRKIVSPAFN------------VEKLKL-LG 549
Query: 193 PLIASVAE---KKTASKADNTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVS 249
P+ + K S +++ LD+ ++ S D + + FG
Sbjct: 550 PIFCECCDDMISKLESVVASSNGSCELDIWPFVQNVSSDVLARAGFGSS---------YE 702
Query: 250 NLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQR-RE 308
F L + RF + +++K K + +I +R +
Sbjct: 703 EGKKVFQLQREMLQLTMTLFKFAFIPGYRFLPTYTNRRMKAIDKEIRGSLMAIINRRLKA 882
Query: 309 IENGVESRKDLLSRFMGALNSHDDEY-------LRDIVVS---FLLAGRDTVASALTGFF 358
I+ G + DLL + + ++ LR++V F LAG++ A L
Sbjct: 883 IKAGEPTNNDLLGILLESNYKESEKSSGGGGMSLREVVEEVKLFYLAGQEANAELLVWTL 1062
Query: 359 ILLSKNPKVEEKIRVELDRVMNPNQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFA 418
+LLSK+P + K R E+ +V E ++ ++ ++ + ES+RL+PPV +++
Sbjct: 1063LLLSKHPDWQAKAREEVFKVFG--NEKPDHDKLGQLKIVSMILQESLRLYPPVVMFARYL 1236
Query: 419 LEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWLKDGVFVPKCPFKYPV 478
+D L D T I G + + + + WG D +EF PER+ + K Y
Sbjct: 1237RKDTKLGDLT-IPAGVEIIVPVSMLHQEKEFWGDDAMEFNPERFSEGVSKATKGKVCYIP 1413
Query: 479 FQAGSRVCLGKELAIVEMKSVVASLVKRFDV 509
F G R+C+G+ ++E K ++ +++ F +
Sbjct: 1414FGWGPRLCIGQNFGLLEAKIALSMILQHFSL 1506
>BP076628
Length = 492
Score = 75.9 bits (185), Expect = 2e-14
Identities = 43/98 (43%), Positives = 58/98 (58%), Gaps = 14/98 (14%)
Frame = -1
Query: 456 EFKPERWLK------------DGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASL 503
EF+PERWL+ FV + F YPVFQAG R+CLGKE+A ++MK +VA +
Sbjct: 489 EFRPERWLEREDTKTNLSPQEKWKFVARDSFSYPVFQAGPRICLGKEMAFLQMKRLVAGI 310
Query: 504 VKRFDVRVVGPN--QEPQFAPGLTASFRGGLPVKIYER 539
+ RF V V P+ +EP F L++ GG PV I+ R
Sbjct: 309 LSRFTV-VPLPHLAEEPNFIAFLSSQMEGGFPVTIHRR 199
>TC13127 weakly similar to UP|O49373 (O49373) Cytochrome P450 - like protein
(Cytochrome P450-like protein), partial (13%)
Length = 495
Score = 74.3 bits (181), Expect = 5e-14
Identities = 38/98 (38%), Positives = 60/98 (60%), Gaps = 2/98 (2%)
Frame = +3
Query: 444 GRMENIWGPDCLEFKPERWLKD-GVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVAS 502
GR E++WG DCLEFKPERW+ + G V +K+ F AG R CLGK+L+ +++K + +
Sbjct: 3 GRSEDVWGKDCLEFKPERWVSERGSIVHAPSYKFISFNAGPRTCLGKDLSFIQLKIIATA 182
Query: 503 LVKRFDVR-VVGPNQEPQFAPGLTASFRGGLPVKIYER 539
+++ + V V G P F+ + + GL V+I +R
Sbjct: 183 VLRNYRVHPVEGHAAIPNFS--IVLLMQDGLKVRITKR 290
>AV406584
Length = 423
Score = 73.9 bits (180), Expect = 6e-14
Identities = 32/55 (58%), Positives = 43/55 (78%)
Frame = +3
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASL 168
T+T +P+N+E+ILKT F+NYPKG ++ T DLLG+GIFN DG +W QRK A+L
Sbjct: 258 TVTCHPKNLEHILKTRFDNYPKGPKWQTAFHDLLGQGIFNSDGETWLMQRKTAAL 422
>TC14281 similar to UP|Q9XHC6 (Q9XHC6) Cytochrome P450 monooxygenaseCYP93D1,
partial (97%)
Length = 1768
Score = 72.8 bits (177), Expect = 1e-13
Identities = 100/433 (23%), Positives = 176/433 (40%), Gaps = 39/433 (9%)
Frame = +3
Query: 115 ITSNPENVEYILKTN---FNNYPKGKQFSTILGDLLGRG----IFNVDGHSWKFQRKMAS 167
+ S+ E + ILKT+ F N P I + L G F G W+F +K+
Sbjct: 291 VVSSAEMAKQILKTSEESFCNRP-----IMIASENLTYGASDYFFIPYGTYWRFLKKLCM 455
Query: 168 LELGS-------VVIRSYAMELVIEEI------------KTRLLPLIASVAEKKTASKAD 208
EL S V +R ++ ++ I + L+ +V T K
Sbjct: 456 TELLSGKTLEHFVSVRENEIQAFLKTILEISKTGEAVVMRQELIRHTNNVISMMTMGKKS 635
Query: 209 NTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALT 268
S+D + +++ ++R + L +F+L + R L
Sbjct: 636 EGSKDEIGELRKVIRE-----------------------IGELLGSFNLGDIIGFMRPLD 746
Query: 269 ASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIE--NGVES--RKDLLSRFM 324
G KK K+ + ++ + +++K+ E G +S +KDL +
Sbjct: 747 LQ------------GYGKKNKDMHRKLDGMMEKVLKEHEEARATEGADSDRKKDLFDILL 890
Query: 325 GALNSH--DDEYLRDIVVSF----LLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRV 378
+ + D++ RD +F LAG + AS L L +NP V +K R E+D V
Sbjct: 891 NLIEADGADNKLTRDSAKAFALDMFLAGTNGPASVLEWSLAELIRNPHVLKKAREEIDTV 1070
Query: 379 MNPNQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTY 438
+ + + YL + E++R+ PP ++ A+ + DG I S++
Sbjct: 1071VG-KERLVKESDIPNLPYLQAVVKETLRMHPPTPIFAREAMRSCQV-DGYDIPANSKIFI 1244
Query: 439 HPYAMGRMENIWGPDCLEFKPERWL---KDGVFVPKCPFKYPVFQAGSRVCLGKELAIVE 495
YA+GR W + + PER+L + V V ++ F +G R C G LA++
Sbjct: 1245SAYAIGRDSQYWENPHV-YDPERFLFTDERKVDVRGQYYQLLPFGSGRRSCPGASLALIV 1421
Query: 496 MKSVVASLVKRFD 508
+++ +ASLV+ FD
Sbjct: 1422IQASLASLVQCFD 1460
>TC8057 weakly similar to UP|Q8W228 (Q8W228) Cytochrome P450, partial (29%)
Length = 1148
Score = 72.0 bits (175), Expect = 2e-13
Identities = 57/236 (24%), Positives = 106/236 (44%), Gaps = 10/236 (4%)
Frame = +2
Query: 283 GSEKKLKEAIKIVNDLANEMIKQRR------EIENG--VESRKDLLSRFMGALNSH--DD 332
G +K K+A + + + ++IK + +NG ++ LLS + + H +
Sbjct: 50 GLVRKFKKAHEEFDQMLEQIIKDHEAPSRLSDQKNGQSIDFTDTLLSHMKQSKDKHIINK 229
Query: 333 EYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQTR 392
L+ I++ ++ DT L ++PK K++ EL V+ + +
Sbjct: 230 TNLKAILLDIIIGSLDTTIMTTDWAMSELLRHPKAMTKLQDELKNVVGMGRVVEE-DDIP 406
Query: 393 EMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGP 452
++ YLN + E+ R+ PP +D+ +G FI K SRV + + +GR IW
Sbjct: 407 KLPYLNMVVKETFRIHPPAPLLVPRECLEDITINGYFIAKKSRVLVNSWTIGRDPKIWSD 586
Query: 453 DCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFD 508
+ +F P+R+ F++ F +G R C G +L + + V+A LV F+
Sbjct: 587 NAEDFYPDRFQNTDTDSHGLHFQFLPFGSGRRRCPGMQLGLTTVPLVLAQLVHCFN 754
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,393,157
Number of Sequences: 28460
Number of extensions: 137011
Number of successful extensions: 1095
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 1045
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1062
length of query: 540
length of database: 4,897,600
effective HSP length: 95
effective length of query: 445
effective length of database: 2,193,900
effective search space: 976285500
effective search space used: 976285500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC127021.9


