

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.12 + phase: 0
(451 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
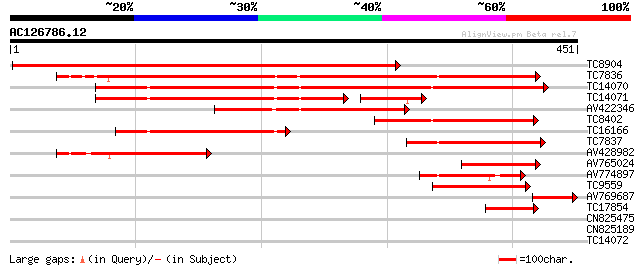
Score E
Sequences producing significant alignments: (bits) Value
TC8904 homologue to UP|G3PB_PEA (P12859) Glyceraldehyde 3-phosph... 573 e-164
TC7836 homologue to UP|G3PA_PEA (P12858) Glyceraldehyde 3-phosph... 563 e-161
TC14070 homologue to UP|G3PC_PEA (P34922) Glyceraldehyde 3-phosp... 301 2e-82
TC14071 similar to UP|G3P_ATRNU (P34783) Glyceraldehyde 3-phosph... 173 5e-44
AV422346 172 8e-44
TC8402 homologue to GB|CAA42103.1|1345501|AMGADPH glycolytic gly... 123 5e-29
TC16166 homologue to UP|G3PC_MAGLI (P26518) Glyceraldehyde 3-pho... 114 3e-26
TC7837 similar to UP|G3PA_PEA (P12858) Glyceraldehyde 3-phosphat... 105 1e-23
AV428982 100 8e-22
AV765024 97 4e-21
AV774897 91 5e-19
TC9559 similar to GB|AAD30223.1|4835756|T8K14 Is a member of the... 78 3e-15
AV769687 58 3e-09
TC17854 homologue to UP|G3PX_HORVU (P26517) Glyceraldehyde 3-pho... 49 2e-06
CN825475 37 0.007
CN825189 35 0.019
TC14072 similar to GB|CAA42103.1|1345501|AMGADPH glycolytic glyc... 28 3.1
>TC8904 homologue to UP|G3PB_PEA (P12859) Glyceraldehyde 3-phosphate
dehydrogenase B, chloroplast precursor (NADP-dependent
glyceraldehydephosphate dehydrogenase subunit B) ,
partial (69%)
Length = 978
Score = 573 bits (1478), Expect = e-164
Identities = 289/309 (93%), Positives = 298/309 (95%)
Frame = +3
Query: 3 THAALASTRIPTSTRFPSKASHSFPTQCASKRLEVAEFSGLKSTSCITYASNARESSFSD 62
THAALASTRIPT+T+F SKASHSFP QC+SKRLEV EFSGL+S+SC+TYA+NARESSF D
Sbjct: 51 THAALASTRIPTNTKFSSKASHSFPAQCSSKRLEVTEFSGLRSSSCVTYANNARESSFFD 230
Query: 63 VVAAQLATKAIGSTAVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSGG 122
VVAAQL KAIGSTAVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSGG
Sbjct: 231 VVAAQLTPKAIGSTAVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSGG 410
Query: 123 VKNASHLLKYDSMLGTFKADVKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEG 182
VKNASHLLKYDSMLGTFKADVKILN TITVDGKPI VV++RDPLKLPWAELGIDIVIEG
Sbjct: 411 VKNASHLLKYDSMLGTFKADVKILNENTITVDGKPIKVVANRDPLKLPWAELGIDIVIEG 590
Query: 183 TGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNEQDYGHEVADIISNASCTTN 242
TGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNE DYGH VADIISNASCTTN
Sbjct: 591 TGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNEGDYGHNVADIISNASCTTN 770
Query: 243 CLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRARAAALNIVPTSTGAAKA 302
CLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRARAAALNIVPTSTGAAKA
Sbjct: 771 CLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDASHRDLRRARAAALNIVPTSTGAAKA 950
Query: 303 VSLVLPQLK 311
VSLVLPQLK
Sbjct: 951 VSLVLPQLK 977
>TC7836 homologue to UP|G3PA_PEA (P12858) Glyceraldehyde 3-phosphate
dehydrogenase A, chloroplast precursor (NADP-dependent
glyceraldehydephosphate dehydrogenase subunit A) ,
partial (89%)
Length = 1513
Score = 563 bits (1450), Expect = e-161
Identities = 293/387 (75%), Positives = 327/387 (83%), Gaps = 2/387 (0%)
Frame = +3
Query: 38 AEFSGLKSTSCITYASNARESSFSDVVAAQLATKAIGSTA--VRGETVAKLKVAINGFGR 95
++FSGL+S + + ++ E F V+A Q T A+GS+ +G AKLKVAINGFGR
Sbjct: 159 SDFSGLRSAN-LPFSRKTSED-FHSVIAFQ--TYAVGSSGGYKKGVVEAKLKVAINGFGR 326
Query: 96 IGRNFLRCWHGRKDSPLEVIVVNDSGGVKNASHLLKYDSMLGTFKADVKILNNETITVDG 155
IGRNFLRCWHGRKDSPL+VI +ND+GGVK ASHLLKYDS LG F ADVK + + I+VDG
Sbjct: 327 IGRNFLRCWHGRKDSPLDVIAINDTGGVKQASHLLKYDSTLGIFGADVKPVGTDGISVDG 506
Query: 156 KPILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIP 215
K I VVS R+P LPW ELGID+VIEGTGVFVD GAGKHIQAGAKKV+ITAP KG DIP
Sbjct: 507 KVIKVVSDRNPANLPWGELGIDLVIEGTGVFVDREGAGKHIQAGAKKVLITAPGKG-DIP 683
Query: 216 TYVVGVNEQDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLL 275
TYVVGVN Y H+ DIISNASCTTNCLAPFVK+LD++FGI+KGTMTTTHSYTGDQRLL
Sbjct: 684 TYVVGVNADAYSHD-DDIISNASCTTNCLAPFVKVLDQKFGIIKGTMTTTHSYTGDQRLL 860
Query: 276 DASHRDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNV 335
DASHRDLRRARAAALNIVPTSTGAAKAV+LVLP LKGKLNGIALRVPTPNVSVVDLVV V
Sbjct: 861 DASHRDLRRARAAALNIVPTSTGAAKAVALVLPTLKGKLNGIALRVPTPNVSVVDLVVQV 1040
Query: 336 AKKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMV 395
KK AE+VNAAFR++A+ L G+L VCD PLVSVDFRCTDVSST+DSSLTMVMGDDMV
Sbjct: 1041TKKTF-AEEVNAAFRESADNELNGILSVCDEPLVSVDFRCTDVSSTVDSSLTMVMGDDMV 1217
Query: 396 KVVAWYDNEWGYSQRVVDLAHLVANKW 422
KV+AWYDNEWGYSQRVVDLA +VAN W
Sbjct: 1218KVIAWYDNEWGYSQRVVDLADIVANNW 1298
>TC14070 homologue to UP|G3PC_PEA (P34922) Glyceraldehyde 3-phosphate
dehydrogenase, cytosolic , complete
Length = 1414
Score = 301 bits (770), Expect = 2e-82
Identities = 171/363 (47%), Positives = 235/363 (64%), Gaps = 3/363 (0%)
Frame = +2
Query: 69 ATKAIGSTAVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSGGVKN-AS 127
A + ST T+ K+K+ INGFGRIGR R R D +E++ VND + +
Sbjct: 8 APRYSSSTLTLSTTMGKIKIGINGFGRIGRLVARVALTRDD--VELVAVNDPFITTDYMT 181
Query: 128 HLLKYDSMLGTFKA-DVKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEGTGVF 186
++ KYDS+ G +K +V + + T+ KP+ V ++R+P ++PW + G DIV+E TGVF
Sbjct: 182 YMFKYDSVHGPWKHHEVTVKDANTLLFGDKPVTVFANRNPEEIPWGQTGADIVVESTGVF 361
Query: 187 VDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNEQDYGHEVADIISNASCTTNCLAP 246
D A H++ GAKKVII+AP+K D P +VVGVNE +Y E+ +IISNASCTTNCLAP
Sbjct: 362 TDKDKAAAHLKGGAKKVIISAPSK--DAPMFVVGVNEHEYKPEL-NIISNASCTTNCLAP 532
Query: 247 FVKILDEEFGIVKGTMTTTHSYTGDQRLLDA-SHRDLRRARAAALNIVPTSTGAAKAVSL 305
K++++ FGIV+G MTT HS T Q+ +D S +D R RAA+ NI+P+STGAAKAV
Sbjct: 533 LAKVINDRFGIVEGLMTTVHSITATQKTVDGPSAKDWRGGRAASFNIIPSSTGAAKAVGK 712
Query: 306 VLPQLKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGPLKGVLDVCD 365
VLP L GKL G+A RVPT +VSVVDL V + +K + + + AA ++ +EG LKG+L +
Sbjct: 713 VLPALNGKLTGMAFRVPTVDVSVVDLTVRL-EKAATYDQIKAAIKEESEGKLKGILGYTE 889
Query: 366 VPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLVANKWPGT 425
+VS DF + SS D+ + + D+ VK+V+WYDNE GYS RVVDL VA G
Sbjct: 890 DDVVSTDFIGDNRSSIFDAKAGIALNDNFVKLVSWYDNEMGYSTRVVDLIVHVAKSL*GV 1069
Query: 426 PQA 428
A
Sbjct: 1070GAA 1078
>TC14071 similar to UP|G3P_ATRNU (P34783) Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) , partial (51%)
Length = 760
Score = 173 bits (439), Expect = 5e-44
Identities = 95/203 (46%), Positives = 133/203 (64%), Gaps = 2/203 (0%)
Frame = +2
Query: 69 ATKAIGSTAVRGETVAKLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSGGVKN-AS 127
+T ST T+ K+K+ INGFGRIGR R R D +E++ VND + +
Sbjct: 56 STATSSSTLTLSTTMGKIKIGINGFGRIGRLVARVALTRDD--VELVAVNDPFITTDYMT 229
Query: 128 HLLKYDSMLGTFKA-DVKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEGTGVF 186
++ KYDS+ G +K +V + + T+ KP+ V ++R+P ++PW + G DIV+E TGVF
Sbjct: 230 YMFKYDSVHGPWKHHEVTVKDANTLLFGDKPVTVFANRNPEEIPWGQTGADIVVESTGVF 409
Query: 187 VDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNEQDYGHEVADIISNASCTTNCLAP 246
D A H++ GAKKVII+AP+K D P +VVGVNE +Y E+ +IISNASCTTNCLAP
Sbjct: 410 TDKDKAAAHLKGGAKKVIISAPSK--DAPMFVVGVNEHEYKPEL-NIISNASCTTNCLAP 580
Query: 247 FVKILDEEFGIVKGTMTTTHSYT 269
K++++ FGIV+G MTT HS T
Sbjct: 581 LAKVINDRFGIVEGLMTTVHSIT 649
Score = 49.3 bits (116), Expect = 1e-06
Identities = 28/57 (49%), Positives = 38/57 (66%), Gaps = 5/57 (8%)
Frame = -2
Query: 280 RDLRRARAAALNIVPTSTGAAKAVSLVLPQLKGKLN----GIALRVPT-PNVSVVDL 331
+D R RAA+ NI+P+STGAAKAV VLP L GKL + ++ T PN+S++ L
Sbjct: 756 KDWRGGRAASFNIIPSSTGAAKAVGKVLPALNGKLTVME*TVVIKPSTMPNLSLITL 586
>AV422346
Length = 465
Score = 172 bits (437), Expect = 8e-44
Identities = 89/156 (57%), Positives = 114/156 (73%), Gaps = 1/156 (0%)
Frame = +2
Query: 164 RDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNE 223
RDP ++PW++ G D VIE +GVF A H++AGAKKVII+AP+ AD P +VVGVNE
Sbjct: 2 RDPAEIPWSDFGADYVIESSGVFTTLEKASSHLKAGAKKVIISAPS--ADAPMFVVGVNE 175
Query: 224 QDYGHEVADIISNASCTTNCLAPFVKILDEEFGIVKGTMTTTHSYTGDQRLLDA-SHRDL 282
+ Y + DI+SNASCTTNCLAP K++ EEFGI++G MTT H+ T Q+ +D S +D
Sbjct: 176 KTYNPSM-DIVSNASCTTNCLAPLAKVVHEEFGILEGLMTTVHATTATQKTVDGPSMKDW 352
Query: 283 RRARAAALNIVPTSTGAAKAVSLVLPQLKGKLNGIA 318
R R AA NI+P+STGAAKAV VLP+L GKL G+A
Sbjct: 353 RGGRGAAQNIIPSSTGAAKAVGKVLPELNGKLTGMA 460
>TC8402 homologue to GB|CAA42103.1|1345501|AMGADPH glycolytic
glyceraldehyde 3-phosphate dehydrogenase {Antirrhinum
majus;} , partial (43%)
Length = 700
Score = 123 bits (309), Expect = 5e-29
Identities = 66/130 (50%), Positives = 87/130 (66%)
Frame = +3
Query: 291 NIVPTSTGAAKAVSLVLPQLKGKLNGIALRVPTPNVSVVDLVVNVAKKGISAEDVNAAFR 350
NI+P+ST AAKAV VLP L GKL G++ RVPT +VSVVDL V + +K S E + AA
Sbjct: 45 NIIPSSTVAAKAVGKVLPSLNGKLTGMSFRVPTVDVSVVDLTVRL-EKAASYEQIKAAIM 221
Query: 351 KAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKVVAWYDNEWGYSQR 410
+ +EG LKG+L + +VS DF SS D+ + + D+ VK+V+WYDNEWGYS R
Sbjct: 222 EESEGKLKGILGYIEEDVVSTDFVGDSRSSIFDAKAGISLNDNFVKLVSWYDNEWGYSTR 401
Query: 411 VVDLAHLVAN 420
VVDL +A+
Sbjct: 402 VVDLIVHIAS 431
>TC16166 homologue to UP|G3PC_MAGLI (P26518) Glyceraldehyde 3-phosphate
dehydrogenase, cytosolic , partial (40%)
Length = 476
Score = 114 bits (285), Expect = 3e-26
Identities = 62/141 (43%), Positives = 92/141 (64%), Gaps = 2/141 (1%)
Frame = +3
Query: 85 KLKVAINGFGRIGRNFLRCWHGRKDSPLEVIVVNDSGGVKN-ASHLLKYDSMLGTFKA-D 142
K+K+ INGFGRIGR R R D +E++ +ND + S++ KYD++ G +K D
Sbjct: 66 KIKIGINGFGRIGRLVARVALQRND--VELVAINDPFITTDYMSYMFKYDTVHGQWKHFD 239
Query: 143 VKILNNETITVDGKPILVVSSRDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKK 202
VK+ +++T+ K + V +R+P ++PW E+G D V+E TGVF D A H++ GAKK
Sbjct: 240 VKVKDSKTLLFGDKAVTVFGTRNPEEIPWGEVGADFVVESTGVFTDKDKAAAHLKGGAKK 419
Query: 203 VIITAPAKGADIPTYVVGVNE 223
V+I+AP+K D P +VVGVNE
Sbjct: 420 VVISAPSK--DAPMFVVGVNE 476
>TC7837 similar to UP|G3PA_PEA (P12858) Glyceraldehyde 3-phosphate
dehydrogenase A, chloroplast precursor (NADP-dependent
glyceraldehydephosphate dehydrogenase subunit A) ,
partial (19%)
Length = 467
Score = 105 bits (263), Expect = 1e-23
Identities = 63/114 (55%), Positives = 76/114 (66%), Gaps = 3/114 (2%)
Frame = +2
Query: 316 GIALRVPTPNVSVVDLVVNVAKKGISAEDVNAAFRKAAEGP-LKGVLDVCDVPLVSVDFR 374
G+ALRVPTP +SVVDLVV V KK AE+VNAA + + L G+L VCD PL SV
Sbjct: 2 GLALRVPTPTLSVVDLVVQVPKKTF-AEEVNAALHERVQTTELHGILSVCDEPLRSVRLL 178
Query: 375 -CTDVSSTIDSSLTMVMGD-DMVKVVAWYDNEWGYSQRVVDLAHLVANKWPGTP 426
+DSSLT+ + +VKV+AWYDNEWGYS RVVDLA LVA+ W +P
Sbjct: 179 GALMCPPPLDSSLTLAPWEMTLVKVIAWYDNEWGYSPRVVDLADLVAHPWK*SP 340
>AV428982
Length = 428
Score = 99.8 bits (247), Expect = 8e-22
Identities = 58/125 (46%), Positives = 78/125 (62%), Gaps = 2/125 (1%)
Frame = +2
Query: 38 AEFSGLKSTSCITYASNARESSFSDVVAAQLATKAIGSTAV--RGETVAKLKVAINGFGR 95
++FSGL+S + + ++ E S A T A+GS+ +G A L VAING+ R
Sbjct: 65 SDFSGLRSAN-LPFSRKTSEDFHS---AIAFHTYAVGSSGGYNKGAEQATLNVAINGYDR 232
Query: 96 IGRNFLRCWHGRKDSPLEVIVVNDSGGVKNASHLLKYDSMLGTFKADVKILNNETITVDG 155
+G+N LRCW GRKD L+VI +ND+ GVK A HL KY+S LG F ADVK + + I+V G
Sbjct: 233 MGKNSLRCWLGRKDIALDVIAINDTAGVKQAYHLFKYESTLGIFGADVKPVGTDGISVHG 412
Query: 156 KPILV 160
K I V
Sbjct: 413 KVIKV 427
>AV765024
Length = 343
Score = 97.4 bits (241), Expect = 4e-21
Identities = 44/63 (69%), Positives = 50/63 (78%)
Frame = -1
Query: 360 VLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLVA 419
+L VCD P V VDFRC+DVSS DSSL MVM D M KV+AWYDNEWG+SQRVVDL ++ A
Sbjct: 343 ILSVCDEPPVPVDFRCSDVSSPADSSLPMVMEDPMPKVIAWYDNEWGHSQRVVDLPYIAA 164
Query: 420 NKW 422
N W
Sbjct: 163 NNW 155
>AV774897
Length = 468
Score = 90.5 bits (223), Expect = 5e-19
Identities = 52/89 (58%), Positives = 62/89 (69%), Gaps = 5/89 (5%)
Frame = -1
Query: 327 SVVDLVVNVAKKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSV-DFRCTDVSS----T 381
SVVDLVV V K AE+VNAAFR++A+ L G+L + D PL++ FRCTDVSS
Sbjct: 468 SVVDLVVQVTSKTF-AEEVNAAFRESADHELNGILSLSDEPLLAQ*TFRCTDVSSHP*LI 292
Query: 382 IDSSLTMVMGDDMVKVVAWYDNEWGYSQR 410
D +GDDMVKV+AWYDNEWGYSQR
Sbjct: 291 FDHGS---LGDDMVKVIAWYDNEWGYSQR 214
>TC9559 similar to GB|AAD30223.1|4835756|T8K14 Is a member of the PF|00044
glyceraldehyde 3-phosphate dehydrogenase family. ESTs
gb|T43985, gb|N38667, gb|N65037, gb|AA713069 and
gb|AI099548 come from this gene. {Arabidopsis
thaliana;}, partial (22%)
Length = 656
Score = 78.2 bits (191), Expect = 3e-15
Identities = 39/78 (50%), Positives = 50/78 (64%)
Frame = +1
Query: 337 KKGISAEDVNAAFRKAAEGPLKGVLDVCDVPLVSVDFRCTDVSSTIDSSLTMVMGDDMVK 396
+K S EDV AA +A+EGPLKG+L D +VS DF SS D+ + + VK
Sbjct: 19 QKNASYEDVKAAI*RASEGPLKGILGYTDEDVVSNDFVGDSRSSIFDAKAGIALSASFVK 198
Query: 397 VVAWYDNEWGYSQRVVDL 414
+V+WYDNEWGYS RV+DL
Sbjct: 199 MVSWYDNEWGYSNRVLDL 252
>AV769687
Length = 342
Score = 58.2 bits (139), Expect = 3e-09
Identities = 25/36 (69%), Positives = 31/36 (85%), Gaps = 1/36 (2%)
Frame = -1
Query: 417 LVANKWPGTPQA-GSGDPLEDFCETNPSTDECKVFE 451
LVA+KWPG A GSGDPLE+FCETNP+ +ECK++E
Sbjct: 342 LVASKWPGAAVAPGSGDPLEEFCETNPADEECKLYE 235
>TC17854 homologue to UP|G3PX_HORVU (P26517) Glyceraldehyde 3-phosphate
dehydrogenase, cytosolic , partial (14%)
Length = 505
Score = 48.9 bits (115), Expect = 2e-06
Identities = 21/42 (50%), Positives = 29/42 (69%)
Frame = +3
Query: 379 SSTIDSSLTMVMGDDMVKVVAWYDNEWGYSQRVVDLAHLVAN 420
SS D+ + + + VK+V+WYDNEWGYS RVVDL +A+
Sbjct: 21 SSIFDAKAGISLNNHFVKLVSWYDNEWGYSSRVVDLIRHMAS 146
>CN825475
Length = 584
Score = 37.0 bits (84), Expect = 0.007
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +3
Query: 432 DPLEDFCETNPSTDECKVFE 451
DPLED+C+ NP T ECK F+
Sbjct: 363 DPLEDYCKDNPETKECKTFD 422
>CN825189
Length = 609
Score = 35.4 bits (80), Expect = 0.019
Identities = 11/20 (55%), Positives = 17/20 (85%)
Frame = +2
Query: 432 DPLEDFCETNPSTDECKVFE 451
DPLE++C+ NP TDEC+ ++
Sbjct: 380 DPLENYCKENPETDECRTYD 439
>TC14072 similar to GB|CAA42103.1|1345501|AMGADPH glycolytic glyceraldehyde
3-phosphate dehydrogenase {Antirrhinum majus;} , partial
(8%)
Length = 374
Score = 28.1 bits (61), Expect = 3.1
Identities = 10/19 (52%), Positives = 15/19 (78%)
Frame = +3
Query: 390 MGDDMVKVVAWYDNEWGYS 408
+ D+ V +V+WYD+E GYS
Sbjct: 6 LNDNFV*LVSWYDHEMGYS 62
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,622,976
Number of Sequences: 28460
Number of extensions: 79226
Number of successful extensions: 445
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 423
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 423
length of query: 451
length of database: 4,897,600
effective HSP length: 93
effective length of query: 358
effective length of database: 2,250,820
effective search space: 805793560
effective search space used: 805793560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC126786.12


