

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
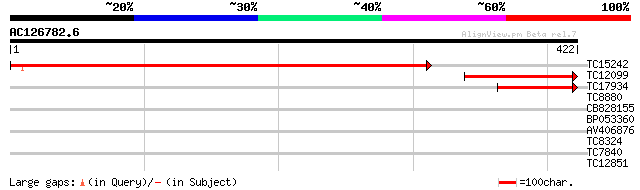
Query= AC126782.6 - phase: 1
(422 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15242 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate synt... 566 e-162
TC12099 similar to PIR|T06667|T06667 argininosuccinate synthase ... 155 2e-38
TC17934 similar to PIR|T06667|T06667 argininosuccinate synthase ... 100 8e-22
TC8880 similar to UP|ST35_ARATH (Q94LW6) Probable sulfate transp... 35 0.030
CB828155 30 0.75
BP053360 29 1.7
AV406876 28 3.7
TC8324 UP|Q8VWY1 (Q8VWY1) Mitochondrial phosphate transporter, c... 27 4.9
TC7840 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced ... 27 6.3
TC12851 UP|Q84TG9 (Q84TG9) Beta-tubulin, partial (34%) 27 6.3
>TC15242 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate synthase,
chloroplast precursor (Citrulline--aspartate ligase) ,
partial (57%)
Length = 1296
Score = 566 bits (1460), Expect = e-162
Identities = 272/316 (86%), Positives = 302/316 (95%), Gaps = 2/316 (0%)
Frame = +2
Query: 1 IKAVLYKD--VEISETKKVAGLKGKLNKVVLAYSGGLDTSVVVPWLRENYGCDVVCFTAD 58
IKAVL D VEISETKK +GL+GKLNKVVLAYSGGLDTSV+VPWLRENYGC+VVCFTAD
Sbjct: 347 IKAVLRNDTDVEISETKKGSGLRGKLNKVVLAYSGGLDTSVIVPWLRENYGCEVVCFTAD 526
Query: 59 LGQGASELEGLEEKAKASGASQLVVKDLKEEFVRDYVFPCLRAGAIYERKYLLGTAIARP 118
+GQG ELEGLEEKAKASGASQLVVKDL+EEFV+DY+FPCLRAGA YERKYLLGTA+ARP
Sbjct: 527 VGQGLKELEGLEEKAKASGASQLVVKDLQEEFVKDYIFPCLRAGATYERKYLLGTAMARP 706
Query: 119 VIAKAMVDIANEVGADAVSHGCTGKGNDQVRFELSFFALNPKLNVVAPWREWDITGREDA 178
VIAKAMVD+A EVGADAVSHGCTGKGNDQVRFEL+FFALNPKLNVVAPWREWDITGREDA
Sbjct: 707 VIAKAMVDVAKEVGADAVSHGCTGKGNDQVRFELTFFALNPKLNVVAPWREWDITGREDA 886
Query: 179 IEYAKKHDVPVPVTKKSIYSRDRNLWHITHEGDVLEDPANEPKKDMYMLTVDPEDAPNEP 238
IEYAKKH++PVPVTKKSIYSRDRNLWH++HEGD+LEDPANEP+KDMYM++VDPEDAP++P
Sbjct: 887 IEYAKKHNIPVPVTKKSIYSRDRNLWHLSHEGDILEDPANEPQKDMYMMSVDPEDAPDKP 1066
Query: 239 EYVEIGFELGIPVSLNGKRLSPGNLVAELNEIGGRHGIGRVDLVEDRIVGMKSRGVYETP 298
EYVEIG E G+PVS+NGKRLSP +L+AELNEIGGRHGIGR+D+VE+R+VGMKSRGVYETP
Sbjct: 1067EYVEIGIEAGLPVSVNGKRLSPASLLAELNEIGGRHGIGRIDMVENRLVGMKSRGVYETP 1246
Query: 299 GGTILFTAARDLETLT 314
GGTILF AAR+LE LT
Sbjct: 1247GGTILFAAARELELLT 1294
>TC12099 similar to PIR|T06667|T06667 argininosuccinate synthase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (16%)
Length = 477
Score = 155 bits (391), Expect = 2e-38
Identities = 71/84 (84%), Positives = 83/84 (98%)
Frame = +3
Query: 339 WFDPLRESMDSFMEKISQTTTGSVSLKLYKGSASVTGRKSPYSLYREDISSFESGEIYNH 398
WFDPLRES+D+FM KI++TTTGSV+LKLYKGSA+VTGRKSPYSLYREDISSF++G++YNH
Sbjct: 3 WFDPLRESLDAFMHKITETTTGSVTLKLYKGSAAVTGRKSPYSLYREDISSFDTGDLYNH 182
Query: 399 ADAVGFIKLYGLPMRIRAMMEQGI 422
ADAVGFIKLYGLPMRIRAMM+QG+
Sbjct: 183 ADAVGFIKLYGLPMRIRAMMQQGL 254
>TC17934 similar to PIR|T06667|T06667 argininosuccinate synthase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (11%)
Length = 564
Score = 99.8 bits (247), Expect = 8e-22
Identities = 46/59 (77%), Positives = 54/59 (90%)
Frame = +2
Query: 364 LKLYKGSASVTGRKSPYSLYREDISSFESGEIYNHADAVGFIKLYGLPMRIRAMMEQGI 422
LKL+KGS VT R+SP+SLYR+DISSFESGEIY+ ADA GFI+LYGLPMR+RAM+EQGI
Sbjct: 2 LKLFKGSVGVTSRQSPFSLYRQDISSFESGEIYDQADAAGFIRLYGLPMRVRAMLEQGI 178
>TC8880 similar to UP|ST35_ARATH (Q94LW6) Probable sulfate transporter 3.5,
partial (30%)
Length = 794
Score = 34.7 bits (78), Expect = 0.030
Identities = 17/51 (33%), Positives = 26/51 (50%)
Frame = -1
Query: 203 LWHITHEGDVLEDPANEPKKDMYMLTVDPEDAPNEPEYVEIGFELGIPVSL 253
+W +GDV+ DP N P + + +L PE E ++ GFE + SL
Sbjct: 296 IWEPLEDGDVVLDPFNNPPRPVLLLLDLPERIIREERLLQPGFEFCVKPSL 144
>CB828155
Length = 542
Score = 30.0 bits (66), Expect = 0.75
Identities = 15/23 (65%), Positives = 19/23 (82%)
Frame = +3
Query: 1 IKAVLYKDVEISETKKVAGLKGK 23
IKAVL +D++I ETK V GL+GK
Sbjct: 477 IKAVL-QDIQIPETKNVTGLRGK 542
>BP053360
Length = 530
Score = 28.9 bits (63), Expect = 1.7
Identities = 16/70 (22%), Positives = 29/70 (40%)
Frame = +3
Query: 85 DLKEEFVRDYVFPCLRAGAIYERKYLLGTAIARPVIAKAMVDIANEVGADAVSHGCTGKG 144
D E+ +RD P +++ K LG ++ +A A + + S CT +G
Sbjct: 111 DDHEDVLRDDSLPVCSTDRVHDVK--LGDSVEESTVASATRSSQTNAASGSSSKACTARG 284
Query: 145 NDQVRFELSF 154
+D F +
Sbjct: 285 SDSADFRSGY 314
>AV406876
Length = 234
Score = 27.7 bits (60), Expect = 3.7
Identities = 18/50 (36%), Positives = 23/50 (46%)
Frame = +3
Query: 118 PVIAKAMVDIANEVGADAVSHGCTGKGNDQVRFELSFFALNPKLNVVAPW 167
P +A D+ VG D S GC G G D VRF + A K++ W
Sbjct: 27 PGVAVNFGDLKGFVGGD--SDGCGGSGEDAVRFVVGELAKLLKVHCDRFW 170
>TC8324 UP|Q8VWY1 (Q8VWY1) Mitochondrial phosphate transporter, complete
Length = 1605
Score = 27.3 bits (59), Expect = 4.9
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Frame = +3
Query: 363 SLKLYKGSASVTGRKSPYSLYREDISSFES--GEIYNHA 399
+L LYKG + GR+ PY++ + +SFE+ IY HA
Sbjct: 771 TLGLYKGLVPLWGRQIPYTMMK--FASFETIVEMIYKHA 881
>TC7840 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced response
protein, complete
Length = 1585
Score = 26.9 bits (58), Expect = 6.3
Identities = 13/32 (40%), Positives = 18/32 (55%)
Frame = -3
Query: 26 KVVLAYSGGLDTSVVVPWLRENYGCDVVCFTA 57
K+V+ L SV PW R + C+V+ FTA
Sbjct: 1133 KLVVVSYVSL*DSVAFPWRRPSLICEVISFTA 1038
>TC12851 UP|Q84TG9 (Q84TG9) Beta-tubulin, partial (34%)
Length = 591
Score = 26.9 bits (58), Expect = 6.3
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 8/69 (11%)
Frame = +3
Query: 123 AMVDIANEVGAD--------AVSHGCTGKGNDQVRFELSFFALNPKLNVVAPWREWDITG 174
A+ DI VG+ A H TG+G D++ F + +N ++ + + + G
Sbjct: 216 AIQDIFKRVGSHFSAMFKRKAFLHWYTGEGMDEMEFTEAESNMNDLVSELQQYEAASVEG 395
Query: 175 REDAIEYAK 183
E+ EYA+
Sbjct: 396 EEEEDEYAE 422
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,966,266
Number of Sequences: 28460
Number of extensions: 69208
Number of successful extensions: 290
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 290
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 290
length of query: 422
length of database: 4,897,600
effective HSP length: 93
effective length of query: 329
effective length of database: 2,250,820
effective search space: 740519780
effective search space used: 740519780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC126782.6


