

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126781.9 + phase: 0
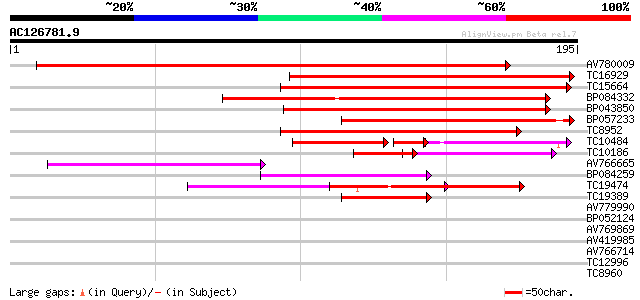
(195 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV780009 186 3e-48
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 98 1e-21
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 96 5e-21
BP084332 94 2e-20
BP043850 91 1e-19
BP057233 77 2e-15
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 74 2e-14
TC10484 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragm... 40 7e-11
TC10186 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprot... 34 9e-07
AV766665 46 4e-06
BP084259 44 2e-05
TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retro... 42 9e-05
TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 41 2e-04
AV779990 32 0.001
BP052124 37 0.003
AV769869 30 0.21
AV419985 27 2.3
AV766714 27 2.3
TC12996 similar to UP|Q84VQ3 (Q84VQ3) NAF specific protein kinas... 26 5.2
TC8960 26 5.2
>AV780009
Length = 529
Score = 186 bits (471), Expect = 3e-48
Identities = 91/163 (55%), Positives = 120/163 (72%)
Frame = -1
Query: 10 EAALRVVRFLKGNPGQGIFFGSKSDLRLHGWCDSDWASCPLTRRSVTGWIVQLGDSPISW 69
+AA RV+R++KG P QG+FF + S L+L + DSDWA CP TRRSVTG+ + LG S ISW
Sbjct: 529 QAATRVLRYVKGAPAQGLFFSADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSLISW 350
Query: 70 KTKKQHTVSRSSAEAEYRSMANTTCELKWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKN 129
+TKKQ TVSRSS+EAEYR++A T CE++WL + L + P+ + CD+Q+A+HIA N
Sbjct: 349 RTKKQTTVSRSSSEAEYRALAATVCEVQWLSYLFQFLKLNVPLPVPLFCDNQSALHIAHN 170
Query: 130 PVFHERTKHIEVDCHFVRNEVLKNNILPVYVSTANQPADIFTK 172
P FHERTKHIE+DCH VR ++ I + +ST +Q ADIFTK
Sbjct: 169 PTFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTK 41
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 97.8 bits (242), Expect = 1e-21
Identities = 45/98 (45%), Positives = 66/98 (66%)
Frame = +1
Query: 97 KWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKNNIL 156
+WL +L +L V ++P +++CD+ +A HIA NPVFHERTKHIE+DCH VR + K I
Sbjct: 1 QWLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIH 180
Query: 157 PVYVSTANQPADIFTKALGKPQFEFLLGKLGIRNLHAP 194
+ +S++ ADI+TKAL F + KLG+ N+ +P
Sbjct: 181 LLPISSSEPLADIYTKALSPQNFHQICAKLGLINICSP 294
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 95.5 bits (236), Expect = 5e-21
Identities = 46/100 (46%), Positives = 65/100 (65%)
Frame = +1
Query: 94 CELKWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKN 153
CEL WL IL +L V P +++CDSQ+A HIA N VFHERTKH+++DCH VR ++
Sbjct: 1 CEL*WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAK 180
Query: 154 NILPVYVSTANQPADIFTKALGKPQFEFLLGKLGIRNLHA 193
+ +S+ +Q ADI TK L F L+ KLG+ N+++
Sbjct: 181 LFHLLPISSVDQTADILTKPLESGPFSHLVSKLGVLNIYS 300
>BP084332
Length = 368
Score = 94.0 bits (232), Expect = 2e-20
Identities = 49/113 (43%), Positives = 73/113 (64%)
Frame = +2
Query: 74 QHTVSRSSAEAEYRSMANTTCELKWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNPVFH 133
Q T++ S+AEAEY S A + ++ W+K L + ++ + I+CD+ AAI ++KNP+ H
Sbjct: 32 QSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESN-IPIYCDNTAAISLSKNPILH 208
Query: 134 ERTKHIEVDCHFVRNEVLKNNILPVYVSTANQPADIFTKALGKPQFEFLLGKL 186
R KHIEV HF+R+ V K +L +V T +Q ADIFTK L + +F F+L L
Sbjct: 209 SRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL 367
>BP043850
Length = 515
Score = 91.3 bits (225), Expect = 1e-19
Identities = 45/92 (48%), Positives = 62/92 (66%)
Frame = -1
Query: 95 ELKWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKNN 154
E+ WL+G+LS LG + +P +H D+ +AI IA NPV+HE T+HIEVDCH VR +
Sbjct: 512 EIIWLRGLLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDCHSVREAYDRRV 333
Query: 155 ILPVYVSTANQPADIFTKALGKPQFEFLLGKL 186
I +VST+ Q ADI TK+L + + FL+ KL
Sbjct: 332 ITLPHVSTSVQIADILTKSLTRQRHNFLVSKL 237
>BP057233
Length = 473
Score = 77.4 bits (189), Expect = 2e-15
Identities = 36/80 (45%), Positives = 54/80 (67%)
Frame = -2
Query: 115 IIHCDSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKNNILPVYVSTANQPADIFTKAL 174
I+ CD+ +A +A NPV H R+KHIE+D H++R++VL+N ++ YV T +Q AD TK L
Sbjct: 448 ILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKPL 269
Query: 175 GKPQFEFLLGKLGIRNLHAP 194
+F L KLG+ +H+P
Sbjct: 268 SHTRFSQLRDKLGV--IHSP 215
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 73.9 bits (180), Expect = 2e-14
Identities = 33/83 (39%), Positives = 58/83 (69%)
Frame = +3
Query: 94 CELKWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKN 153
CE WL L++L + ++I+CD+++A+H+A N VFH+RT++IE+DCH V +VL
Sbjct: 18 CEALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFG 197
Query: 154 NILPVYVSTANQPADIFTKALGK 176
+ ++V +++Q AD+FTK + +
Sbjct: 198 ILHLLHVPSSDQVADVFTKTISQ 266
>TC10484 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (20%)
Length = 479
Score = 40.0 bits (92), Expect(3) = 7e-11
Identities = 16/33 (48%), Positives = 23/33 (69%)
Frame = +3
Query: 98 WLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNP 130
W+ ILS +G+ PM + CD+QAA+HI+ NP
Sbjct: 3 WVGQILSEMGIERISPMPLWCDNQAALHISSNP 101
Score = 32.3 bits (72), Expect(3) = 7e-11
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +2
Query: 133 HERTKHIEVDCH 144
HERTKHIEVDCH
Sbjct: 113 HERTKHIEVDCH 148
Score = 28.5 bits (62), Expect(3) = 7e-11
Identities = 21/53 (39%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Frame = +1
Query: 144 HFVRNEVLKNNILPVYVSTANQPADIFTKALGKPQFEFLLGKLG---IRNLHA 193
+FVRN + I YV TA D+FTKAL + +F+ +L I LHA
Sbjct: 151 YFVRN-FNRTLISTSYVRTATHLPDVFTKALNGVRNDFICNQLRH*VISMLHA 306
>TC10186 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (4%)
Length = 528
Score = 33.9 bits (76), Expect(2) = 9e-07
Identities = 19/53 (35%), Positives = 27/53 (50%)
Frame = +3
Query: 136 TKHIEVDCHFVRNEVLKNNILPVYVSTANQPADIFTKALGKPQFEFLLGKLGI 188
T +IE HF+R++V K I + T Q ADI TK L +F + L +
Sbjct: 63 TLNIETKYHFLRDQVTKGKISLKHCGTDLQVADIMTKGLKTERFRNMRAMLNV 221
Score = 33.9 bits (76), Expect(2) = 9e-07
Identities = 14/22 (63%), Positives = 19/22 (85%)
Frame = +2
Query: 119 DSQAAIHIAKNPVFHERTKHIE 140
D+++AI +AKNPV H R+KHIE
Sbjct: 5 DNKSAIDLAKNPVSHGRSKHIE 70
>AV766665
Length = 601
Score = 46.2 bits (108), Expect = 4e-06
Identities = 22/75 (29%), Positives = 43/75 (57%)
Frame = +3
Query: 14 RVVRFLKGNPGQGIFFGSKSDLRLHGWCDSDWASCPLTRRSVTGWIVQLGDSPISWKTKK 73
R+ +LK N +G F + + G+ +D+ + R S G+ + L + ++W++K+
Sbjct: 372 RIF*YLKANSRRGPLFQKEGKSSMDGFTYADYLGSIVDRLSTMGYYMFLSGNLVTWRSKQ 551
Query: 74 QHTVSRSSAEAEYRS 88
Q+ ++RSS EAE R+
Sbjct: 552 QNIIARSSGEAELRA 596
>BP084259
Length = 385
Score = 43.5 bits (101), Expect = 2e-05
Identities = 20/59 (33%), Positives = 31/59 (51%)
Frame = -3
Query: 87 RSMANTTCELKWLKGILSNLGVVHDKPMIIHCDSQAAIHIAKNPVFHERTKHIEVDCHF 145
R++ +T E WL+ +L L K I ++ A ++ N VFH KH+ +DCHF
Sbjct: 179 RTIGSTAAEFLWLQQLLKELQFSLPKVPCIFSNNINATYVCLNLVFHSHMKHVALDCHF 3
>TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retroelement pol
polyprotein, partial (3%)
Length = 517
Score = 41.6 bits (96), Expect = 9e-05
Identities = 26/68 (38%), Positives = 41/68 (60%), Gaps = 1/68 (1%)
Frame = -2
Query: 111 DKPMIIHC-DSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKNNILPVYVSTANQPADI 169
D +I H D++AA+H+ K F R KHIE+D HF+ + L +I +V++ +Q D+
Sbjct: 381 DVSLITHT*DNKAALHMPKI*-FSMRAKHIEIDFHFL*DRRLYLDISTRFVNSNDQLTDV 205
Query: 170 FTKALGKP 177
FTK+ P
Sbjct: 204 FTKSWRGP 181
Score = 39.3 bits (90), Expect = 5e-04
Identities = 31/90 (34%), Positives = 44/90 (48%)
Frame = -1
Query: 62 LGDSPISWKTKKQHTVSRSSAEAEYRSMANTTCELKWLKGILSNLGVVHDKPMIIHCDSQ 121
L D+ ISWK+K+ V+RS AEAE+R MA T + G V D H
Sbjct: 517 LSDNLISWKSKETIIVARSRAEAEFRVMATATXSK*FASKT*RFGGCVTDN---THLRQ* 347
Query: 122 AAIHIAKNPVFHERTKHIEVDCHFVRNEVL 151
+ A+N VFHE H + F+R +++
Sbjct: 346 GSPTHAQNLVFHEGQTH*D*LPFFIR*KIV 257
>TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (6%)
Length = 498
Score = 40.8 bits (94), Expect = 2e-04
Identities = 14/31 (45%), Positives = 24/31 (77%)
Frame = +2
Query: 115 IIHCDSQAAIHIAKNPVFHERTKHIEVDCHF 145
+++CD+Q AI++ +N FH R+KHI+V H+
Sbjct: 170 VLYCDNQGAIYLGQNSTFHSRSKHIDVRYHW 262
Score = 38.1 bits (87), Expect = 0.001
Identities = 33/125 (26%), Positives = 62/125 (49%), Gaps = 3/125 (2%)
Frame = +3
Query: 59 IVQLGDSPISWKTKKQHTVSRSSAEAEYRSMANTTCELKWLKGILSNLGVVHDKPMIIHC 118
+V ++W ++ Q V+ S+AEAE+ + EL W+K L N H P I+ C
Sbjct: 3 LVTFAGGAVAWPSRLQKCVALSTAEAEFIAATEACHELLWMKNFLQN-AWFHSHP-ILCC 176
Query: 119 --DSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKNNILPV-YVSTANQPADIFTKALG 175
++A +A+ +F + + +R +VL + +L + + T + AD+ TK+L
Sbjct: 177 IVITKALFTLARILLFIQDPSTLMFVIIGLR-DVLNSKLLELEKIHTDDDGADMMTKSLP 353
Query: 176 KPQFE 180
+ + E
Sbjct: 354 REKLE 368
>AV779990
Length = 542
Score = 32.0 bits (71), Expect(2) = 0.001
Identities = 12/20 (60%), Positives = 16/20 (80%)
Frame = +3
Query: 119 DSQAAIHIAKNPVFHERTKH 138
D+ AA+ I+ NP+ HERTKH
Sbjct: 312 DNHAALDISSNPISHERTKH 371
Score = 25.0 bits (53), Expect(2) = 0.001
Identities = 11/24 (45%), Positives = 17/24 (70%)
Frame = +1
Query: 138 HIEVDCHFVRNEVLKNNILPVYVS 161
+IEVDCHF+ E L+ N++ +S
Sbjct: 367 NIEVDCHFI-CEKLEQNLISTSIS 435
>BP052124
Length = 467
Score = 36.6 bits (83), Expect = 0.003
Identities = 19/51 (37%), Positives = 28/51 (54%)
Frame = -2
Query: 118 CDSQAAIHIAKNPVFHERTKHIEVDCHFVRNEVLKNNILPVYVSTANQPAD 168
CD+ +A+ +A P+ H R H EVDC + + E L ++ YV PAD
Sbjct: 451 CDNNSALTLAPRPI*HSRPVHFEVDCPYPK-EKLGTGLITAYV-IFRTPAD 305
>AV769869
Length = 289
Score = 30.4 bits (67), Expect = 0.21
Identities = 15/30 (50%), Positives = 18/30 (60%)
Frame = -3
Query: 165 QPADIFTKALGKPQFEFLLGKLGIRNLHAP 194
Q ADI TK L F L KLG+ N+H+P
Sbjct: 281 QLADILTKPLTLGPFRSLFFKLGMHNIHSP 192
>AV419985
Length = 408
Score = 26.9 bits (58), Expect = 2.3
Identities = 17/55 (30%), Positives = 24/55 (42%), Gaps = 2/55 (3%)
Frame = +1
Query: 3 QPRTEHWEAA--LRVVRFLKGNPGQGIFFGSKSDLRLHGWCDSDWASCPLTRRSV 55
+PR W + + L G P G+ GS RLHG W CPL + ++
Sbjct: 193 RPRHPQWAPTPPMPLPPHLAGRPRSGLLGGSHPCCRLHGRLRRRW--CPLPQAAL 351
>AV766714
Length = 545
Score = 26.9 bits (58), Expect = 2.3
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = -3
Query: 134 ERTKHIEVDCHFVRNE 149
ERTKHI + CHFV E
Sbjct: 255 ERTKHIGLICHFVLKE 208
>TC12996 similar to UP|Q84VQ3 (Q84VQ3) NAF specific protein kinase family,
partial (17%)
Length = 523
Score = 25.8 bits (55), Expect = 5.2
Identities = 11/34 (32%), Positives = 19/34 (55%)
Frame = +3
Query: 45 WASCPLTRRSVTGWIVQLGDSPISWKTKKQHTVS 78
W+S T+ S W + G ++ K+KKQ+ V+
Sbjct: 174 WSSISSTKNSRHAWKMLFGKQKMTCKSKKQNDVN 275
>TC8960
Length = 556
Score = 25.8 bits (55), Expect = 5.2
Identities = 14/53 (26%), Positives = 22/53 (41%)
Frame = -1
Query: 125 HIAKNPVFHERTKHIEVDCHFVRNEVLKNNILPVYVSTANQPADIFTKALGKP 177
H+ NP+ + ++ CHF R P ++ +NQP D K P
Sbjct: 493 HLESNPLERKMYQNTPKRCHFCRQN------SPYTLALSNQPIDFTPKQKKNP 353
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.135 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,034,147
Number of Sequences: 28460
Number of extensions: 60150
Number of successful extensions: 322
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 321
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 321
length of query: 195
length of database: 4,897,600
effective HSP length: 85
effective length of query: 110
effective length of database: 2,478,500
effective search space: 272635000
effective search space used: 272635000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC126781.9


