

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
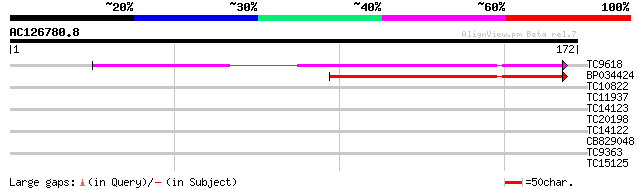
Query= AC126780.8 - phase: 0 /pseudo
(172 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9618 similar to UP|Q41050 (Q41050) Core protein, complete 83 3e-17
BP034424 73 2e-14
TC10822 30 0.22
TC11937 weakly similar to UP|AAS49102 (AAS49102) At1g72640, part... 28 1.1
TC14123 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, parti... 27 1.9
TC20198 similar to UP|Q9ZPU0 (Q9ZPU0) At2g13980 protein, partial... 25 5.5
TC14122 similar to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, c... 25 5.5
CB829048 25 7.2
TC9363 similar to UP|IM17_ARATH (Q9SP35) Mitochondrial import in... 25 7.2
TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enz... 25 9.4
>TC9618 similar to UP|Q41050 (Q41050) Core protein, complete
Length = 763
Score = 82.8 bits (203), Expect = 3e-17
Identities = 49/144 (34%), Positives = 73/144 (50%)
Frame = +2
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+P +N + F+K + A +A + + Y + +G+ N
Sbjct: 209 DMGNPFLNLTVDGFLKIGAVAATRAAAEDTYHIIKKGSISSN------------------ 334
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
D Q KE WG AGLY G+ YG++ RG DWKN+ + GA+TGA ++ S
Sbjct: 335 --DFQKTLKKMCKEGAYWGTVAGLYVGMEYGVERIRGRRDWKNAMLGGAVTGALVSAASG 508
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
+ S ++IA AITGAAI+TAA +
Sbjct: 509 DKS-DKIAIDAITGAAIATAAEFI 577
>BP034424
Length = 531
Score = 73.2 bits (178), Expect = 2e-14
Identities = 36/72 (50%), Positives = 49/72 (68%)
Frame = -1
Query: 98 KESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAI 157
KE WG AGLY G+ YG++ RG DWKN+ + GA+TGA ++ S + S ++IA AI
Sbjct: 336 KEGAYWGTVAGLYVGMEYGVERIRGRRDWKNAMLGGAVTGALVSAASGDKS-DKIAIDAI 160
Query: 158 TGAAISTAANLL 169
TGAAI+TAA +
Sbjct: 159 TGAAIATAAEFI 124
>TC10822
Length = 619
Score = 30.0 bits (66), Expect = 0.22
Identities = 14/44 (31%), Positives = 21/44 (46%)
Frame = +3
Query: 97 GKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAAL 140
G + G+Y G+ ++ RG D N AV G + GAA+
Sbjct: 285 GNHGLTFAAIGGVYIGVEQLVQNFRGKRDLVNGAVGGFVAGAAI 416
>TC11937 weakly similar to UP|AAS49102 (AAS49102) At1g72640, partial (60%)
Length = 774
Score = 27.7 bits (60), Expect = 1.1
Identities = 11/17 (64%), Positives = 13/17 (75%)
Frame = +2
Query: 19 FNKGGLFDFGHPLVNRI 35
F KGGL DF H LVN++
Sbjct: 695 FEKGGLCDFSHLLVNKL 745
>TC14123 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, partial (89%)
Length = 1130
Score = 26.9 bits (58), Expect = 1.9
Identities = 11/29 (37%), Positives = 18/29 (61%)
Frame = +2
Query: 17 SDFNKGGLFDFGHPLVNRIAESFVKAAGI 45
+D GGL D +PLV+ + +S K+ G+
Sbjct: 509 TDRKSGGLGDLNYPLVSDVTKSISKSYGV 595
>TC20198 similar to UP|Q9ZPU0 (Q9ZPU0) At2g13980 protein, partial (13%)
Length = 630
Score = 25.4 bits (54), Expect = 5.5
Identities = 12/19 (63%), Positives = 12/19 (63%)
Frame = -2
Query: 142 CTSDNTSHEQIAQCAITGA 160
C DN S EQI Q AIT A
Sbjct: 308 CLQDNRSQEQIHQNAITSA 252
>TC14122 similar to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, complete
Length = 1107
Score = 25.4 bits (54), Expect = 5.5
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = +1
Query: 22 GGLFDFGHPLVNRIAESFVKAAGI 45
GGL D +PLV+ + +S K+ G+
Sbjct: 550 GGLGDLNYPLVSDVTKSISKSYGV 621
>CB829048
Length = 509
Score = 25.0 bits (53), Expect = 7.2
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = -2
Query: 3 SNSNLETRTLLDELSDFNKGGLFDFGHPL 31
SN+N++ L+++ + N LF+FG PL
Sbjct: 478 SNANMDILCPLEDMPETNPLPLFNFGIPL 392
>TC9363 similar to UP|IM17_ARATH (Q9SP35) Mitochondrial import inner
membrane translocase subunit TIM17, partial (68%)
Length = 1085
Score = 25.0 bits (53), Expect = 7.2
Identities = 18/51 (35%), Positives = 22/51 (42%)
Frame = +3
Query: 103 WGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIA 153
WG GL+S M R D NS AGA TG L+ T+ + A
Sbjct: 240 WG---GLFSSFDCTMVYLRQKEDPWNSIFAGAATGGFLSMRQGLTASTRAA 383
>TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enzyme 12,
partial (48%)
Length = 1164
Score = 24.6 bits (52), Expect = 9.4
Identities = 14/43 (32%), Positives = 20/43 (45%)
Frame = -1
Query: 124 HDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAA 166
HDW+ + V + G A + +CAI A ISTA+
Sbjct: 972 HDWRAAHVGDIMQGNAPV---------NVTRCAIANAHISTAS 871
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,332,234
Number of Sequences: 28460
Number of extensions: 23343
Number of successful extensions: 111
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 109
length of query: 172
length of database: 4,897,600
effective HSP length: 84
effective length of query: 88
effective length of database: 2,506,960
effective search space: 220612480
effective search space used: 220612480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC126780.8


