

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.3 + phase: 0
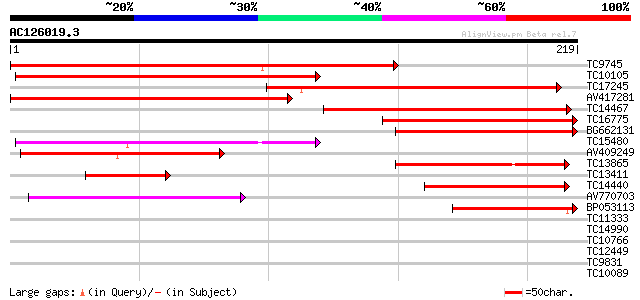
(219 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9745 similar to UP|V727_ARATH (Q9M376) Vesicle-associated memb... 203 2e-53
TC10105 similar to UP|Q7X9C5 (Q7X9C5) Synptobrevin-related prote... 191 6e-50
TC17245 similar to UP|V721_ARATH (Q9ZTW3) Vesicle-associated mem... 177 2e-45
AV417281 172 3e-44
TC14467 similar to UP|Q84SZ1 (Q84SZ1) Synaptobrevin-like protein... 154 1e-38
TC16775 similar to GB|AAA56991.1|600710|ATHSAR1 formerly called ... 111 1e-25
BG662131 85 8e-18
TC15480 similar to UP|V712_ARATH (Q9SIQ9) Vesicle-associated mem... 77 2e-15
AV409249 68 1e-12
TC13865 similar to UP|V714_ARATH (Q9FMR5) Vesicle-associated mem... 62 1e-10
TC13411 similar to UP|V727_ARATH (Q9M376) Vesicle-associated mem... 57 2e-09
TC14440 similar to UP|V711_ARATH (O49377) Vesicle-associated mem... 53 5e-08
AV770703 51 1e-07
BP053113 49 9e-07
TC11333 similar to GB|AAO42855.1|28416649|BT004609 At4g10170 {Ar... 32 0.065
TC14990 UP|Q8S2V3 (Q8S2V3) Small G-protein ROP9, complete 31 0.19
TC10766 similar to UP|Q9SML5 (Q9SML5) Knolle, partial (39%) 29 0.72
TC12449 similar to GB|AAO42852.1|28416643|BT004606 At5g59380 {Ar... 28 0.94
TC9831 similar to UP|Q84VZ3 (Q84VZ3) At5g52990, partial (16%) 28 1.2
TC10089 28 1.6
>TC9745 similar to UP|V727_ARATH (Q9M376) Vesicle-associated membrane
protein 727 (AtVAMP727), partial (70%)
Length = 721
Score = 203 bits (517), Expect = 2e-53
Identities = 95/169 (56%), Positives = 130/169 (76%), Gaps = 19/169 (11%)
Frame = +3
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
M Q+ LIYSFVA+GTV+LAE++ +TGNF+TIA+QCLQKLP++++++TY+CDGHTF+FL+D
Sbjct: 213 MSQRGLIYSFVAKGTVVLAEHTQYTGNFSTIAVQCLQKLPSNSSKYTYSCDGHTFNFLLD 392
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGG-------------------RATTA 101
NGF + VVA ESVGR +P FLER+KDDF +RYG + +
Sbjct: 393 NGFVFLVVADESVGRSVPFVFLERVKDDFMQRYGASIKNASDHPLADDDDDDDLFQDRFS 572
Query: 102 TAKSLNKEFGPKLKEQMQYCVEHPEEVSKLAKVKAQVSEVKGVMMENID 150
A +L++EFGP LKE MQYC+ HPEE+SKL+K+KAQ++EVKG+MM+NI+
Sbjct: 573 IAYNLDREFGPALKEHMQYCLNHPEEMSKLSKLKAQITEVKGIMMDNIE 719
>TC10105 similar to UP|Q7X9C5 (Q7X9C5) Synptobrevin-related protein
(Fragment), partial (59%)
Length = 549
Score = 191 bits (486), Expect = 6e-50
Identities = 86/118 (72%), Positives = 106/118 (88%)
Frame = +1
Query: 3 QQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNG 62
Q+ LIY+FV+RGTVILAE+++F+GNF +IA QCLQKLP+++N+FTYNCD HTF+FLVDNG
Sbjct: 196 QKPLIYAFVSRGTVILAEFTEFSGNFNSIAFQCLQKLPSTSNKFTYNCDAHTFNFLVDNG 375
Query: 63 FTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
+TYC+VA ESVGRQ+P+AFLER+KDDF +YG G+A TA A SLNKEFGPKLKE MQY
Sbjct: 376 YTYCIVADESVGRQVPMAFLERVKDDFVSKYGDGKAATAPANSLNKEFGPKLKEHMQY 549
>TC17245 similar to UP|V721_ARATH (Q9ZTW3) Vesicle-associated membrane
protein 721 (AtVAMP721) (v-SNARE synaptobrevin 7B)
(AtVAMP7B), partial (51%)
Length = 627
Score = 177 bits (448), Expect = 2e-45
Identities = 96/140 (68%), Positives = 104/140 (73%), Gaps = 26/140 (18%)
Frame = +2
Query: 100 TATAKSLNKEFG--------------------------PKLKEQMQYCVEHPEEVSKLAK 133
TA SLNKEFG PKLKE MQYCV+HPEEVSKLAK
Sbjct: 2 TAAPNSLNKEFGYLFEFVLRLA*L*LCS*L*MIFALCRPKLKEHMQYCVDHPEEVSKLAK 181
Query: 134 VKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWYQNMK 193
VKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENL QAQDFR GT++RRKMW QNMK
Sbjct: 182 VKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLHHQAQDFRNSGTKIRRKMWLQNMK 361
Query: 194 IKLIVLAIIIALILIIVLSV 213
IKLIVLAI+IALILIIVL +
Sbjct: 362 IKLIVLAILIALILIIVLPI 421
>AV417281
Length = 423
Score = 172 bits (437), Expect = 3e-44
Identities = 83/109 (76%), Positives = 94/109 (86%)
Frame = +3
Query: 1 MGQQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVD 60
MG+QSLIYSFVARGTVILAE++DF GNF +ALQ LQKLP+SN +FTYN DGHTF++LV
Sbjct: 96 MGEQSLIYSFVARGTVILAEHTDFVGNFGDVALQFLQKLPSSNTKFTYNTDGHTFNYLVH 275
Query: 61 NGFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKE 109
NGFTYCVVAVES GRQI +AFLERIK+DF KRY GG+A TA K LNKE
Sbjct: 276 NGFTYCVVAVESTGRQIAMAFLERIKEDFTKRYDGGKAATAAPKGLNKE 422
>TC14467 similar to UP|Q84SZ1 (Q84SZ1) Synaptobrevin-like protein, partial
(44%)
Length = 602
Score = 154 bits (389), Expect = 1e-38
Identities = 78/96 (81%), Positives = 85/96 (88%)
Frame = +2
Query: 122 VEHPEEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGT 181
V+HPEE+SKLAKVKAQVSEVKGVMMENI+KV+DRGEKIE+LVDKTENL QAQDFR GT
Sbjct: 2 VDHPEEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLHHQAQDFRTSGT 181
Query: 182 QLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGF 217
+RRKMW NMKIKLIVL I+IALILIIVLS GF
Sbjct: 182 SIRRKMWLPNMKIKLIVLGILIALILIIVLSATRGF 289
>TC16775 similar to GB|AAA56991.1|600710|ATHSAR1 formerly called HAT24;
synaptobrevin-related protein {Arabidopsis thaliana;} ,
partial (35%)
Length = 567
Score = 111 bits (277), Expect = 1e-25
Identities = 51/75 (68%), Positives = 64/75 (85%)
Frame = +2
Query: 145 MMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIA 204
MMENI+KV+DRGEKIE+LVD+T+ LRSQA DFR GTQ+RRKMW+ NMKIKLIV II+
Sbjct: 2 MMENIEKVLDRGEKIELLVDETDTLRSQAPDFRPPGTQIRRKMWFPNMKIKLIVAGIIVL 181
Query: 205 LILIIVLSVCHGFSC 219
LILI+++S+C +C
Sbjct: 182 LILILIISICGSLNC 226
>BG662131
Length = 484
Score = 85.1 bits (209), Expect = 8e-18
Identities = 39/70 (55%), Positives = 55/70 (77%)
Frame = +3
Query: 150 DKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILII 209
+ V+DRGEKIE+LVDKTE+L+ QA F++QG QLRRKMW QN+++KL+V I+ L++I+
Sbjct: 15 ENVLDRGEKIELLVDKTESLQFQADSFQRQGRQLRRKMWLQNLQMKLMVGGGILILVIIL 194
Query: 210 VLSVCHGFSC 219
+ C GF C
Sbjct: 195 WVIACGGFKC 224
>TC15480 similar to UP|V712_ARATH (Q9SIQ9) Vesicle-associated membrane
protein 712 (AtVAMP712), partial (53%)
Length = 482
Score = 77.4 bits (189), Expect = 2e-15
Identities = 45/119 (37%), Positives = 67/119 (55%), Gaps = 1/119 (0%)
Frame = +1
Query: 3 QQSLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNN-RFTYNCDGHTFSFLVDN 61
+ ++Y+ VARG+V LAE+S N +TIA Q L K+P +N+ +Y+ D F +
Sbjct: 124 RMGILYALVARGSVALAEFSGTATNASTIAKQILDKIPGNNDTHVSYSQDRFIFHLKRTD 303
Query: 62 GFTYCVVAVESVGRQIPIAFLERIKDDFNKRYGGGRATTATAKSLNKEFGPKLKEQMQY 120
G T +A ++ GR+IP AFLE I F + YG +A +N EF L +QM+Y
Sbjct: 304 GLTVLCMADDNAGRRIPFAFLEEIHQRFVRTYGRA-VFSAQPYGMNDEFSRVLSQQMEY 477
>AV409249
Length = 286
Score = 68.2 bits (165), Expect = 1e-12
Identities = 33/80 (41%), Positives = 55/80 (68%), Gaps = 1/80 (1%)
Frame = +2
Query: 5 SLIYSFVARGTVILAEYSDFTGNFTTIALQCLQKLP-ASNNRFTYNCDGHTFSFLVDNGF 63
+++Y+ VARGTV+LAE+S TGN +A + ++KLP S++R ++ D + F L +G
Sbjct: 44 AILYALVARGTVVLAEFSAVTGNTGAVARRIIEKLPEESDSRLCFSQDRYIFHILRSDGL 223
Query: 64 TYCVVAVESVGRQIPIAFLE 83
TY +A ++ GR+IP ++LE
Sbjct: 224 TYLCMANDTFGRRIPFSYLE 283
>TC13865 similar to UP|V714_ARATH (Q9FMR5) Vesicle-associated membrane
protein 714 (AtVAMP714), partial (34%)
Length = 543
Score = 61.6 bits (148), Expect = 1e-10
Identities = 27/67 (40%), Positives = 47/67 (69%)
Frame = +2
Query: 150 DKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILII 209
+K+++RG++IE+LVDKT ++ + FR+Q +LRR +W +N K+ L +L +I LIL
Sbjct: 2 EKILERGDRIELLVDKTATMQDNSFHFRKQSKRLRRALWMKNFKL-LALLTCLIVLILYF 178
Query: 210 VLSVCHG 216
+++ C G
Sbjct: 179 IIAACCG 199
>TC13411 similar to UP|V727_ARATH (Q9M376) Vesicle-associated membrane
protein 727 (AtVAMP727), partial (14%)
Length = 575
Score = 57.0 bits (136), Expect = 2e-09
Identities = 22/33 (66%), Positives = 30/33 (90%)
Frame = +1
Query: 30 TIALQCLQKLPASNNRFTYNCDGHTFSFLVDNG 62
TI +QC QKLP++ +++TY+CDGHTF+FLVDNG
Sbjct: 232 TIVVQCFQKLPSNCSKYTYSCDGHTFNFLVDNG 330
>TC14440 similar to UP|V711_ARATH (O49377) Vesicle-associated membrane
protein 711 (AtVAMP711) (v-SNARE synaptobrevin 7C)
(AtVAMP7C), partial (28%)
Length = 704
Score = 52.8 bits (125), Expect = 5e-08
Identities = 20/56 (35%), Positives = 38/56 (67%)
Frame = +1
Query: 161 VLVDKTENLRSQAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHG 216
+LVDKT N++ FR+Q + R +W++N+K+ + ++ II+ +I +++ VCHG
Sbjct: 1 LLVDKTANMQGNTFRFRKQARRFRSTVWWRNVKLTIALILIILVIIYVVLAFVCHG 168
>AV770703
Length = 539
Score = 51.2 bits (121), Expect = 1e-07
Identities = 22/84 (26%), Positives = 42/84 (49%)
Frame = +2
Query: 8 YSFVARGTVILAEYSDFTGNFTTIALQCLQKLPASNNRFTYNCDGHTFSFLVDNGFTYCV 67
Y V++G +L YS +A CL+ P + + ++ FL+++G+ Y
Sbjct: 92 YCCVSKGNHVLYAYSGAGQEVENVAALCLESAPPFHRWYFETAGKRSYGFLMEDGYVYFT 271
Query: 68 VAVESVGRQIPIAFLERIKDDFNK 91
+ E +G + + FLE ++D+F K
Sbjct: 272 IVDEGLGNSVVLRFLEHVRDEFRK 343
>BP053113
Length = 464
Score = 48.5 bits (114), Expect = 9e-07
Identities = 25/49 (51%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Frame = -2
Query: 172 QAQDFRQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVC-HGFSC 219
QA FR QG LRRKMW+QN+K+KLIV I+ I C GF+C
Sbjct: 229 QAPVFRGQGDPLRRKMWFQNLKMKLIVFGILFCFISCSCSFQCVRGFNC 83
>TC11333 similar to GB|AAO42855.1|28416649|BT004609 At4g10170 {Arabidopsis
thaliana;}, partial (14%)
Length = 487
Score = 32.3 bits (72), Expect = 0.065
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = +1
Query: 177 RQQGTQLRRKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
R +RRK W++ ++I L + A + ++ +I L +C G SC
Sbjct: 43 RSAPRNIRRK-WWRQVRIVLAIDAAVCIILFVIWLVICRGISC 168
>TC14990 UP|Q8S2V3 (Q8S2V3) Small G-protein ROP9, complete
Length = 1029
Score = 30.8 bits (68), Expect = 0.19
Identities = 18/71 (25%), Positives = 33/71 (46%), Gaps = 10/71 (14%)
Frame = +3
Query: 114 LKEQMQYCVEHP----------EEVSKLAKVKAQVSEVKGVMMENIDKVIDRGEKIEVLV 163
L++ Q+C++HP EE+ KL A + E EN+ V D ++ +
Sbjct: 591 LRDDKQFCIDHPGAVPITTAQGEELRKLINAPAYI-ECSSKTQENVKAVFDAAIRVVLQP 767
Query: 164 DKTENLRSQAQ 174
K + +++AQ
Sbjct: 768 PKQKKKKNKAQ 800
>TC10766 similar to UP|Q9SML5 (Q9SML5) Knolle, partial (39%)
Length = 592
Score = 28.9 bits (63), Expect = 0.72
Identities = 23/82 (28%), Positives = 42/82 (51%), Gaps = 1/82 (1%)
Frame = +3
Query: 133 KVKAQVSEVKGVMMENIDKVIDRGEKIEVLVDKTENLRSQAQDFRQQGTQ-LRRKMWYQN 191
+V+ + E+ V ++ V +GEK+ D E+ + + + GT+ L+ YQ
Sbjct: 111 EVEKSLLELHQVFLDMAVMVEAQGEKM----DDIEHHVLHSSHYVKDGTKNLQTAKMYQK 278
Query: 192 MKIKLIVLAIIIALILIIVLSV 213
K + + III LILI+V+ +
Sbjct: 279 SSRKWMCIGIIILLILILVIVI 344
>TC12449 similar to GB|AAO42852.1|28416643|BT004606 At5g59380 {Arabidopsis
thaliana;}, partial (7%)
Length = 873
Score = 28.5 bits (62), Expect = 0.94
Identities = 10/28 (35%), Positives = 18/28 (63%)
Frame = -1
Query: 188 WYQNMKIKLIVLAIIIALILIIVLSVCH 215
W++ +KL+ ++I L L+ +SVCH
Sbjct: 132 WHKPHNLKLVSARVLIQLFLLNTISVCH 49
>TC9831 similar to UP|Q84VZ3 (Q84VZ3) At5g52990, partial (16%)
Length = 543
Score = 28.1 bits (61), Expect = 1.2
Identities = 10/35 (28%), Positives = 22/35 (62%)
Frame = +3
Query: 185 RKMWYQNMKIKLIVLAIIIALILIIVLSVCHGFSC 219
+ +W +++ + L++ + A++ +I L VC GF C
Sbjct: 93 KHVWKKHVWVVLLLDLFVCAVLFVIWLWVCSGFKC 197
>TC10089
Length = 836
Score = 27.7 bits (60), Expect = 1.6
Identities = 14/42 (33%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Frame = +2
Query: 176 FRQQGTQLRRKMWYQNMKIKLIVLAIIIALI---LIIVLSVC 214
FRQ G W Q + KL++L +++ L+ L+++L VC
Sbjct: 20 FRQLGFVQFSSPWLQRIPRKLLLLHLVVVLVVVFLLLLLPVC 145
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,077,080
Number of Sequences: 28460
Number of extensions: 33460
Number of successful extensions: 219
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 216
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 217
length of query: 219
length of database: 4,897,600
effective HSP length: 87
effective length of query: 132
effective length of database: 2,421,580
effective search space: 319648560
effective search space used: 319648560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 53 (25.0 bits)
Medicago: description of AC126019.3


