

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.13 - phase: 0
(367 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
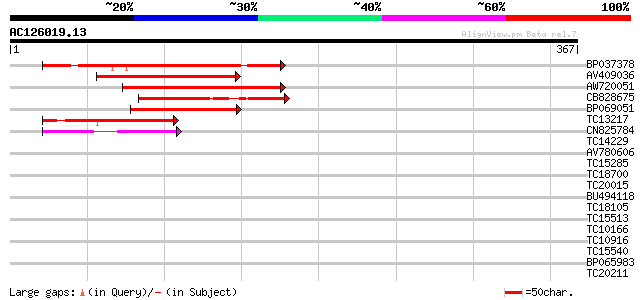
Score E
Sequences producing significant alignments: (bits) Value
BP037378 178 1e-45
AV409036 124 2e-29
AW720051 112 7e-26
CB828675 112 1e-25
BP069051 108 2e-24
TC13217 similar to UP|Q9M4G1 (Q9M4G1) Dof zinc finger protein, p... 100 5e-22
CN825784 51 3e-07
TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%) 39 0.002
AV780606 34 0.044
TC15285 homologue to PIR|T10586|T10586 small nuclear ribonucleop... 33 0.075
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 33 0.098
TC20015 32 0.13
BU494118 32 0.13
TC18105 UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (5%) 31 0.28
TC15513 homologue to UP|O61085 (O61085) Coronin binding protein,... 30 0.63
TC10166 weakly similar to UP|Q8W0T8 (Q8W0T8) Gb protein, partial... 30 0.83
TC10916 similar to UP|Q94KS0 (Q94KS0) Histidine-containing phosp... 29 1.1
TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fra... 29 1.1
BP065983 29 1.1
TC20211 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, p... 29 1.1
>BP037378
Length = 546
Score = 178 bits (451), Expect = 1e-45
Identities = 96/166 (57%), Positives = 113/166 (67%), Gaps = 9/166 (5%)
Frame = +1
Query: 22 QQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQSQPH-GGGSTGSIR--PGSMSDRAR---- 74
QQ N HQ GN GS+ L P QQ Q PH GGG GSI P +D+AR
Sbjct: 31 QQPNQHQANGNDGSLHHLL----PPLAQQQQHAPHVGGGGGGSISSSPSLTADQARAQAQ 198
Query: 75 --MANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGG 132
+A M P ALKCPRCESTNTKFCYFNNYSLSQPRHFCKTC+RYWTRGGALRNVPVGG
Sbjct: 199 AQLAKMAPPPEALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCKRYWTRGGALRNVPVGG 378
Query: 133 GFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSS 178
GFRRNK+ KS+++ +ST + SD+Q+ S S+TN + N+ S+
Sbjct: 379 GFRRNKKHKKSNSSSSST---SESDKQSLSNSTTNELNIPTNMTST 507
>AV409036
Length = 429
Score = 124 bits (311), Expect = 2e-29
Identities = 58/94 (61%), Positives = 68/94 (71%), Gaps = 1/94 (1%)
Frame = +1
Query: 57 GGGSTGSIRPGSMSDRARMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCR 116
GGG G R +DR + ALKCPRC+S NTKFCY+NNY+LSQPRHFCK CR
Sbjct: 91 GGGGGGGSRFFGGADRRLRPHHQQNNQALKCPRCDSINTKFCYYNNYNLSQPRHFCKNCR 270
Query: 117 RYWTRGGALRNVPVGGGFRRN-KRSSKSSNNGNS 149
RYWT+GG LRNVPVGGG R++ KRSSK N+ +S
Sbjct: 271 RYWTKGGVLRNVPVGGGCRKSTKRSSKPKNSSSS 372
>AW720051
Length = 564
Score = 112 bits (281), Expect = 7e-26
Identities = 51/105 (48%), Positives = 66/105 (62%)
Frame = +1
Query: 74 RMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGG 133
R P LKCPRCES NTKFCY+NNY+LSQPRHFCK CRRYWT+GG LRN+ VG G
Sbjct: 103 RKVPRPHHHQILKCPRCESLNTKFCYYNNYNLSQPRHFCKGCRRYWTKGGLLRNITVGSG 282
Query: 134 FRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSS 178
R+ KRS + ++ P + Q+ S S+ S + + ++
Sbjct: 283 SRKPKRSINKNTVPSAPPPPPPPEPQSSSGGSSTLTESTSKMFAA 417
>CB828675
Length = 552
Score = 112 bits (279), Expect = 1e-25
Identities = 54/98 (55%), Positives = 70/98 (71%)
Frame = +3
Query: 84 ALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNKRSSKS 143
ALKCPRC+STNTKFCY+NNY+ SQPRH+C+ C+R+WT+GG LRNVPV GG R+NK+ K
Sbjct: 273 ALKCPRCDSTNTKFCYYNNYNKSQPRHYCRACKRHWTKGGTLRNVPV-GGVRKNKKIKK- 446
Query: 144 SNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADI 181
ST SP SS T S S+T + N++ +A +
Sbjct: 447 -----STTSP-SSGTTTTSISTTTTSNGKNDVDFNAPL 542
>BP069051
Length = 359
Score = 108 bits (269), Expect = 2e-24
Identities = 46/72 (63%), Positives = 56/72 (76%)
Frame = -2
Query: 79 PMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGFRRNK 138
P E A+ CPRC S NT+FCY+NNYSL+QPR+FC TCRRYWT GG+LR++PVGGG R+NK
Sbjct: 313 PQTEEAVNCPRCHSINTQFCYYNNYSLTQPRYFC*TCRRYWTEGGSLRHIPVGGGSRKNK 134
Query: 139 RSSKSSNNGNST 150
S S + ST
Sbjct: 133 HRSNSVISTPST 98
>TC13217 similar to UP|Q9M4G1 (Q9M4G1) Dof zinc finger protein, partial
(15%)
Length = 422
Score = 100 bits (248), Expect = 5e-22
Identities = 52/95 (54%), Positives = 62/95 (64%), Gaps = 7/95 (7%)
Frame = +1
Query: 22 QQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQSQP-------HGGGSTGSIRPGSMSDRAR 74
QQQ ++QQ G + Q + PPPQ S GGG GSIRPGSM+DRAR
Sbjct: 148 QQQPNNQQVG----IFPQQNSPRPPPPQPPPSTDIVGGGGGTGGGYQGSIRPGSMADRAR 315
Query: 75 MANMPMPETALKCPRCESTNTKFCYFNNYSLSQPR 109
+A + P+ LKCPRCESTNTKFCY+NNY+LSQPR
Sbjct: 316 LAKIHQPDATLKCPRCESTNTKFCYYNNYNLSQPR 420
>CN825784
Length = 548
Score = 50.8 bits (120), Expect = 3e-07
Identities = 24/90 (26%), Positives = 41/90 (44%)
Frame = +1
Query: 22 QQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMSDRARMANMPMP 81
+ +N PG + ++ +T + QS P + + P
Sbjct: 319 ETKNSSTSPGAVVNPETPSIEEETAKSKSEQSDP--------------AQSQEKTTLKKP 456
Query: 82 ETALKCPRCESTNTKFCYFNNYSLSQPRHF 111
+ + CPRC S +TKFCY++NY ++QPR+F
Sbjct: 457 DKVIPCPRCNSMDTKFCYYHNYHVNQPRYF 546
>TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%)
Length = 1175
Score = 38.5 bits (88), Expect = 0.002
Identities = 47/212 (22%), Positives = 74/212 (34%), Gaps = 11/212 (5%)
Frame = +1
Query: 16 LCCLLLQQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSIRPGSMSDRA-- 73
LC ++L Q ++ N + L+ ++ QSQS P S+ D+A
Sbjct: 229 LCLIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFP 408
Query: 74 -------RMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALR 126
A+ + + P + N + H C C + + G AL
Sbjct: 409 SYQALGGHKASHRKSSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKSFPTGQAL- 585
Query: 127 NVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTP 186
GG R + KSSN+ NS + SS + TS+ S+ + G
Sbjct: 586 ----GGHKRCHYEGGKSSNSTNSNVH--------ANTSSAVTTTSDGGAASAHSLRGFDL 729
Query: 187 QMSS--LRFMTPLHHNFSNPNDFVGGGDLGLN 216
+ + F TPL +GGGD N
Sbjct: 730 NLPAPLTEFWTPLG---------IGGGDSRTN 798
>AV780606
Length = 585
Score = 33.9 bits (76), Expect = 0.044
Identities = 34/127 (26%), Positives = 53/127 (40%), Gaps = 8/127 (6%)
Frame = -1
Query: 56 HGGGSTGSIRP----GSMSDRARMANMPMPETALKCPRCESTNTKFCYFNNYSLSQPRHF 111
H G ST S + GS +D+ + ET+ C C T++K S PR
Sbjct: 534 HKGQSTSSKKQDGANGSGTDQESGQDDSQAETS--CTHC-GTSSKSTPMMRRGPSGPRSL 364
Query: 112 CKTCRRYWTRGGALRNVPVGGGFRRNKRSS----KSSNNGNSTKSPASSDRQTGSASSTN 167
C C +W GALR++ +RN+ S + + GN S+D +T + N
Sbjct: 363 CNACGLFWANRGALRDLS-----KRNQEHSLVQVEQVDGGN------SADCRTAMPALNN 217
Query: 168 SITSNNN 174
+ + N
Sbjct: 216 VVAFSEN 196
>TC15285 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (61%)
Length = 587
Score = 33.1 bits (74), Expect = 0.075
Identities = 22/50 (44%), Positives = 26/50 (52%), Gaps = 5/50 (10%)
Frame = -3
Query: 45 TPP----PQQSQSQP-HGGGSTGSIRPGSMSDRARMANMPMPETALKCPR 89
TPP PQ+S QP H GG G G + R R A+ P PE A + PR
Sbjct: 357 TPPAEADPQRSWKQPPHRGGERGR-GCGDLRGRRRRASCPWPEEACETPR 211
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 32.7 bits (73), Expect = 0.098
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 8/56 (14%)
Frame = -3
Query: 17 CCL-------LLQQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQS-QPHGGGSTGSI 64
CCL L+ QQ H+Q S+ Q L + P QQ ++ QPH GSTGSI
Sbjct: 226 CCL**WLQNNLIHQQEQHKQ-NQYSSLHQGLPHIEIPANQQCKTLQPHQ*GSTGSI 62
>TC20015
Length = 599
Score = 32.3 bits (72), Expect = 0.13
Identities = 20/72 (27%), Positives = 28/72 (38%)
Frame = +2
Query: 129 PVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQM 188
P GG +NN N T S TG +T S T +N I + + PQ+
Sbjct: 344 PQPGGLIDQSHHMLYNNNNNGTNSRKRGREVTGIGHTTTSTTPSNGIMNQFSLQSQPPQL 523
Query: 189 SSLRFMTPLHHN 200
+L + H N
Sbjct: 524 INLSQLHNHHQN 559
>BU494118
Length = 222
Score = 32.3 bits (72), Expect = 0.13
Identities = 19/52 (36%), Positives = 31/52 (59%)
Frame = +3
Query: 127 NVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSS 178
+VP+GG R RSS +S + P S+DR TG+ +S+++ S ++ SS
Sbjct: 27 SVPIGGALDRRSRSS------SSWRIP-SADRSTGTTTSSSATASTSSARSS 161
>TC18105 UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (5%)
Length = 550
Score = 31.2 bits (69), Expect = 0.28
Identities = 20/67 (29%), Positives = 27/67 (39%)
Frame = +2
Query: 8 FSNFLPFCLCCLLLQQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSIRPG 67
FS F+P L L Q NHH P + ++ Q PPP S + G + +I
Sbjct: 356 FSLFIP-----LFLFQFNHHSHPFSLRHKRNEMRPIQLPPPSTSATGSPGAAANNNINRD 520
Query: 68 SMSDRAR 74
R R
Sbjct: 521 QRPQRRR 541
>TC15513 homologue to UP|O61085 (O61085) Coronin binding protein, partial
(5%)
Length = 1029
Score = 30.0 bits (66), Expect = 0.63
Identities = 19/48 (39%), Positives = 23/48 (47%)
Frame = +3
Query: 17 CCLLLQQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQSQPHGGGSTGSI 64
C +L +Q HHQQP + S QQ PPP S P +T SI
Sbjct: 159 CAGVLHRQIHHQQPHHHRSHHQQ------PPPPLSLPNPATTTTTSSI 284
>TC10166 weakly similar to UP|Q8W0T8 (Q8W0T8) Gb protein, partial (5%)
Length = 980
Score = 29.6 bits (65), Expect = 0.83
Identities = 20/71 (28%), Positives = 31/71 (43%)
Frame = +3
Query: 120 TRGGALRNVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSA 179
T GGA R+ G R SS +NN ++ SS + AS + S+ S+++ S
Sbjct: 756 TAGGASRSASSAGSGDRTGPSSGDNNNSSNALPSDSSIKGARVASGSGSVLSSSDYTGSG 935
Query: 180 DILGLTPQMSS 190
+ SS
Sbjct: 936 PVSSPAVHFSS 968
>TC10916 similar to UP|Q94KS0 (Q94KS0) Histidine-containing phosphotransfer
protein, partial (90%)
Length = 781
Score = 29.3 bits (64), Expect = 1.1
Identities = 29/108 (26%), Positives = 42/108 (38%), Gaps = 15/108 (13%)
Frame = -3
Query: 8 FSNFLPFCLCCLLLQQQNHHQ-----------QPGNGGSVSQQLLQTQTPPPQQSQSQPH 56
F F+P CL C L + HQ Q + SQ L + +TP Q+Q H
Sbjct: 386 FLPFVPQCLSCYPLTDEREHQLF*SQHLSALEQKRDL*EASQNLQKRETPLQPQTQGCSH 207
Query: 57 GGGS--TGSIRPGSMSDRARMANMPMPETALKC--PRCESTNTKFCYF 100
G + + PGS P+T + C P S++ + C F
Sbjct: 206 LGAAEVAETDHPGS-----------PPQTGI*CSPPTVSSSDPRPCCF 96
>TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fragment),
partial (6%)
Length = 806
Score = 29.3 bits (64), Expect = 1.1
Identities = 18/53 (33%), Positives = 24/53 (44%), Gaps = 1/53 (1%)
Frame = +3
Query: 126 RNVPVGGGFRRNKRSSKSSNNGNSTKSPASSDRQTGSASSTNSIT-SNNNIHS 177
RN + N + SSNN +S+ S+ SS NS+ SNNN S
Sbjct: 12 RNCSSSSNYNSNHHNLPSSNNNSSSNFHKSNSSSFPRCSSNNSLNFSNNNSRS 170
>BP065983
Length = 356
Score = 29.3 bits (64), Expect = 1.1
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = -3
Query: 15 CLCCLLLQQQNHHQQPGNGGSVSQQLLQTQTPPPQQSQ 52
C C + Q N HQ N SQ L ++ +PPP SQ
Sbjct: 348 CYSCRVYFQLNRHQYRHNRRRRSQFLHRSPSPPPSPSQ 235
>TC20211 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, partial
(67%)
Length = 623
Score = 29.3 bits (64), Expect = 1.1
Identities = 21/57 (36%), Positives = 31/57 (53%)
Frame = +2
Query: 141 SKSSNNGNSTKSPASSDRQTGSASSTNSITSNNNIHSSADILGLTPQMSSLRFMTPL 197
S + +G+ T SP S SST+S++ + ++S LTP+ SSLR TPL
Sbjct: 239 SATPASGSPTSSPGSP-----KTSSTSSLSPPSALNSLPAP*MLTPKSSSLRSGTPL 394
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.128 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,650,239
Number of Sequences: 28460
Number of extensions: 124016
Number of successful extensions: 939
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 916
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 936
length of query: 367
length of database: 4,897,600
effective HSP length: 92
effective length of query: 275
effective length of database: 2,279,280
effective search space: 626802000
effective search space used: 626802000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC126019.13


