

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126018.10 + phase: 0
(72 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
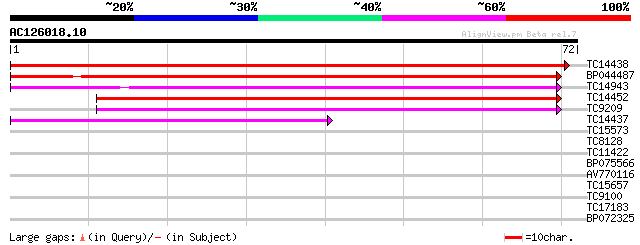
Score E
Sequences producing significant alignments: (bits) Value
TC14438 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteo... 103 8e-24
BP044487 54 6e-09
TC14943 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protea... 49 1e-07
TC14452 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease... 44 8e-06
TC9209 homologue to UP|Q7X9M5 (Q7X9M5) ATP-dependent Clp proteas... 39 1e-04
TC14437 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteo... 37 7e-04
TC15573 similar to UP|O82421 (O82421) ATP-dependent Clp protease... 32 0.023
TC8128 homologue to UP|TBA3_ELEIN (O22349) Tubulin alpha-3 chain... 26 1.6
TC11422 UP|NIA_LOTJA (P39869) Nitrate reductase [NADH] (NR) , c... 25 2.8
BP075566 25 3.6
AV770116 24 6.2
TC15657 similar to UP|Q40977 (Q40977) Monodehydroascorbate reduc... 23 8.0
TC9100 similar to UP|Q8W544 (Q8W544) NADH-ubiquinone oxidoreduct... 23 8.0
TC17183 weakly similar to UP|Q95RC5 (Q95RC5) LD44506p (CG5745-PA... 23 8.0
BP072325 23 8.0
>TC14438 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteolytic
subunit (Endopeptidase Clp) , complete
Length = 620
Score = 103 bits (256), Expect = 8e-24
Identities = 49/71 (69%), Positives = 58/71 (81%)
Frame = +3
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQPA+S YE Q GE +LEAEELLK+R+TIT +Y QRTGK W + DMERD+FMSA E
Sbjct: 399 MIHQPASSFYEAQTGEFILEAEELLKLRETITRVYVQRTGKPLWLVSEDMERDVFMSAAE 578
Query: 61 AEAHGIVDTVA 71
A+A+GIVD VA
Sbjct: 579 AQAYGIVDLVA 611
>BP044487
Length = 533
Score = 53.9 bits (128), Expect = 6e-09
Identities = 24/70 (34%), Positives = 46/70 (65%)
Frame = -1
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP+ Y GQ + + ++++++ + ++YA+ TG++ I ++M+RD FM+ EE
Sbjct: 506 MIHQPSGG-YSGQAKDIAIHTKQIVRVWDLLNDLYAKHTGQSIELIQKNMDRDYFMTPEE 330
Query: 61 AEAHGIVDTV 70
A+ G++D V
Sbjct: 329 AKEFGLIDEV 300
>TC14943 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (ATP-dependent Clp protease proteolytic subunit
ClpP5) (Endopeptidase Clp) , partial (42%)
Length = 639
Score = 49.3 bits (116), Expect = 1e-07
Identities = 25/70 (35%), Positives = 42/70 (59%)
Frame = +1
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEE 60
MIHQP GQ + ++A E+L + + + TG++ +I +D +RD FMSA+E
Sbjct: 130 MIHQPLGGAQGGQT-DIDIQANEMLHHKANLNGYLSYHTGQSLEKINQDTDRDFFMSAKE 306
Query: 61 AEAHGIVDTV 70
A+ +G++D V
Sbjct: 307 AKEYGLIDGV 336
>TC14452 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease-like
protein (ATP-dependent Clp protease proteolytic
subunit) (Endopeptidase Clp) (At5g45390) , partial
(36%)
Length = 621
Score = 43.5 bits (101), Expect = 8e-06
Identities = 20/59 (33%), Positives = 37/59 (61%)
Frame = +2
Query: 12 GQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAHGIVDTV 70
GQ + ++A+E++ + +T I + TG+ Q+ +D++RD +MS EA +GI+D V
Sbjct: 11 GQAIDVEIQAKEVMHNKNNVTRIISGFTGRTFEQVQKDIDRDRYMSPIEAVEYGIIDGV 187
>TC9209 homologue to UP|Q7X9M5 (Q7X9M5) ATP-dependent Clp protease
proteolytic subunit (Fragment), complete
Length = 595
Score = 39.3 bits (90), Expect = 1e-04
Identities = 20/59 (33%), Positives = 35/59 (58%)
Frame = +2
Query: 12 GQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAHGIVDTV 70
GQ + EA ELL++R + N A +TG+ +I +D+ R +A+EA +G++D +
Sbjct: 140 GQADDIQNEANELLRIRDYLFNELATKTGQPVEKITQDLSRMKRFNAQEALEYGLIDRI 316
>TC14437 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteolytic
subunit (Endopeptidase Clp) , partial (58%)
Length = 1013
Score = 37.0 bits (84), Expect = 7e-04
Identities = 17/41 (41%), Positives = 23/41 (55%)
Frame = +3
Query: 1 MIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGK 41
MIHQPA+S YE Q GE + + + + +Y QRT K
Sbjct: 888 MIHQPASSFYEAQTGEFISGSGRTPETTRNYHRVYVQRTAK 1010
>TC15573 similar to UP|O82421 (O82421) ATP-dependent Clp protease
proteolytic subunit (Endopeptidase Clp) (Fragment) ,
partial (25%)
Length = 567
Score = 32.0 bits (71), Expect = 0.023
Identities = 16/32 (50%), Positives = 21/32 (65%)
Frame = +1
Query: 39 TGKASWQIYRDMERDLFMSAEEAEAHGIVDTV 70
TGK QI D +RD FM+ EA+ +G+VD V
Sbjct: 10 TGKPEEQIELDTDRDNFMNPWEAKEYGLVDGV 105
>TC8128 homologue to UP|TBA3_ELEIN (O22349) Tubulin alpha-3 chain (Alpha-3
tubulin), complete
Length = 1615
Score = 25.8 bits (55), Expect = 1.6
Identities = 15/49 (30%), Positives = 21/49 (42%)
Frame = -3
Query: 14 VGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAE 62
VG L +EL + + A ++IY D RD+F A AE
Sbjct: 410 VGGVFLAGDELFGVEELAVGAGADFVDDGGFEIYEDGARDVFTGAGFAE 264
>TC11422 UP|NIA_LOTJA (P39869) Nitrate reductase [NADH] (NR) , complete
Length = 2932
Score = 25.0 bits (53), Expect = 2.8
Identities = 16/56 (28%), Positives = 27/56 (47%), Gaps = 2/56 (3%)
Frame = -1
Query: 3 HQPATSLYEGQVGEC--MLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFM 56
+ P + L+ C + +E L+++ K++ Q T ASWQIY E F+
Sbjct: 2440 YHPRSCLHRPHTS*CDPLGPSESLVQLGKSV*FQSHQLTWLASWQIYASREPKSFL 2273
>BP075566
Length = 253
Score = 24.6 bits (52), Expect = 3.6
Identities = 10/22 (45%), Positives = 13/22 (58%)
Frame = -3
Query: 41 KASWQIYRDMERDLFMSAEEAE 62
K W I+++ E DLFM A E
Sbjct: 245 KEDWYIHQNPEPDLFMKANXCE 180
>AV770116
Length = 206
Score = 23.9 bits (50), Expect = 6.2
Identities = 8/20 (40%), Positives = 15/20 (75%)
Frame = +2
Query: 15 GECMLEAEELLKMRKTITNI 34
G+C +EAEE+ +++ TN+
Sbjct: 113 GKCNVEAEEMARLKSKGTNV 172
>TC15657 similar to UP|Q40977 (Q40977) Monodehydroascorbate reductase ,
partial (48%)
Length = 915
Score = 23.5 bits (49), Expect = 8.0
Identities = 11/33 (33%), Positives = 18/33 (54%)
Frame = +2
Query: 4 QPATSLYEGQVGECMLEAEELLKMRKTITNIYA 36
+P TSL++GQV E + + + N+YA
Sbjct: 128 RPLTSLFKGQVEEDKGGIKTDSSFKSNVPNVYA 226
>TC9100 similar to UP|Q8W544 (Q8W544) NADH-ubiquinone oxidoreductase
(Fragment), partial (97%)
Length = 712
Score = 23.5 bits (49), Expect = 8.0
Identities = 8/18 (44%), Positives = 12/18 (66%)
Frame = +3
Query: 36 AQRTGKASWQIYRDMERD 53
A+R G A W + +D +RD
Sbjct: 108 AEREGGADWAVQQDADRD 161
>TC17183 weakly similar to UP|Q95RC5 (Q95RC5) LD44506p (CG5745-PA), partial
(6%)
Length = 678
Score = 23.5 bits (49), Expect = 8.0
Identities = 8/17 (47%), Positives = 14/17 (82%)
Frame = -2
Query: 27 MRKTITNIYAQRTGKAS 43
+R+ + NIY +++GKAS
Sbjct: 245 VRRKLANIYTRKSGKAS 195
>BP072325
Length = 153
Score = 23.5 bits (49), Expect = 8.0
Identities = 9/26 (34%), Positives = 14/26 (53%)
Frame = +2
Query: 20 EAEELLKMRKTITNIYAQRTGKASWQ 45
E + K RKT+ + + +G SWQ
Sbjct: 56 ERRQTNKQRKTVNDYGTESSGHVSWQ 133
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.129 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 946,139
Number of Sequences: 28460
Number of extensions: 7173
Number of successful extensions: 41
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 41
length of query: 72
length of database: 4,897,600
effective HSP length: 48
effective length of query: 24
effective length of database: 3,531,520
effective search space: 84756480
effective search space used: 84756480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 48 (23.1 bits)
Medicago: description of AC126018.10


