

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126010.8 - phase: 0
(727 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
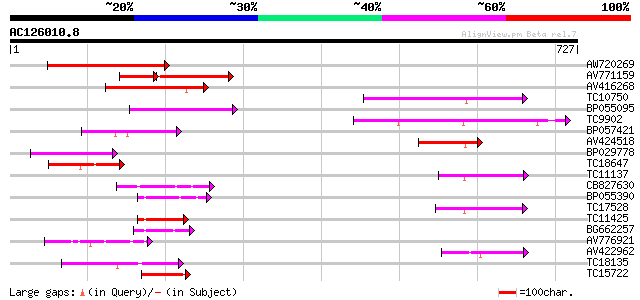
Score E
Sequences producing significant alignments: (bits) Value
AW720269 143 1e-34
AV771159 78 8e-24
AV416268 104 6e-23
TC10750 similar to UP|AAP80385 (AAP80385) ABC transporter, parti... 89 2e-18
BP055095 80 9e-16
TC9902 similar to UP|BAD07483 (BAD07483) PDR-type ABC transporte... 79 3e-15
BP057421 71 5e-13
AV424518 70 2e-12
BP029778 69 2e-12
TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partia... 69 3e-12
TC11137 similar to GB|CAD59563.1|27368811|OSA535041 PDR-like ABC... 64 1e-10
CB827630 55 5e-08
BP055390 54 7e-08
TC17528 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Ar... 52 3e-07
TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proeti... 50 2e-06
BG662257 50 2e-06
AV776921 49 4e-06
AV422962 47 1e-05
TC18135 homologue to UP|Q9MAY4 (Q9MAY4) ABC transporter homolog,... 46 2e-05
TC15722 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transpor... 45 3e-05
>AW720269
Length = 524
Score = 143 bits (360), Expect = 1e-34
Identities = 74/159 (46%), Positives = 110/159 (68%), Gaps = 2/159 (1%)
Frame = +3
Query: 49 VDLLHDITGYAPKGCITAVMGPSGAGKSTLLDGLAGRIASGSLKGK-VSLDGNSVNA-SL 106
V++L ++ A I AV+GPSG GKSTLL +AGR+ K +S++ + + + +
Sbjct: 48 VNILKSVSFVARSSEIVAVVGPSGTGKSTLLRIIAGRVKDRDFDPKTISINDHPMTSPAQ 227
Query: 107 IKRTSAYIMQEDRLFPMLTVYETLMFAADFRLGPLSAVDKRQRVEKLIEQLGLSSSRNTY 166
+++ ++ QED L P+LTV ETL+F+A FRL ++ D+ RVE L+++LGL +++
Sbjct: 228 LRKICGFVAQEDNLLPLLTVKETLLFSAKFRLKEMTPNDREMRVESLMQELGLFHVSDSF 407
Query: 167 IGDEGTRGVSGGERRRVSIGVDIIHGPSLLFLDEPTSGL 205
+GDE RG+SGGER+RVSIGVD+IH P +L LDEPTSGL
Sbjct: 408 VGDEENRGISGGERKRVSIGVDMIHNPPILLLDEPTSGL 524
>AV771159
Length = 486
Score = 77.8 bits (190), Expect(2) = 8e-24
Identities = 40/98 (40%), Positives = 61/98 (61%)
Frame = -3
Query: 189 IIHGPSLLFLDEPTSGLDSTSALSVIEKLHDIARNGSTVILTIHQPSSRIQLLLDHLIIL 248
+IH PSLL LDEPTSGLDST+AL ++ + +A G TV+ TIHQPSSR+ + D +++L
Sbjct: 337 LIH-PSLLLLDEPTSGLDSTTALRILNTIPRLATGGRTVVTTIHQPSSRLYYMFDKVVLL 161
Query: 249 ARGQLMFQGSLKDVGHHLNRMGRKIPKGENPIENLIDV 286
+ G ++ G + + +G NP + L+D+
Sbjct: 160 SEGCPIYYGPASTALEYFSSVGFSTCVTVNPADLLLDL 47
Score = 50.1 bits (118), Expect(2) = 8e-24
Identities = 25/51 (49%), Positives = 35/51 (68%)
Frame = -2
Query: 141 LSAVDKRQRVEKLIEQLGLSSSRNTYIGDEGTRGVSGGERRRVSIGVDIIH 191
L+ +K Q VE++I +LGLS R++ IG RG+SG E+RRVSIG + H
Sbjct: 485 LTRDEKVQHVERVITELGLSGCRSSMIGGPLLRGISGAEKRRVSIGQETAH 333
>AV416268
Length = 424
Score = 104 bits (259), Expect = 6e-23
Identities = 56/136 (41%), Positives = 90/136 (66%), Gaps = 4/136 (2%)
Frame = +3
Query: 124 LTVYETLMFAADFRL-GPLSAVDKRQRVEKLIEQLGLSSSRNTYIGDEGTRGVSGGERRR 182
LTV E + ++A +L +S +KR+R + I+++GL + NT IG G++GVSGG++RR
Sbjct: 12 LTVGEAVYYSAHLQLPDSMSKSEKRERADFTIKEMGLQDAINTRIGGWGSKGVSGGQKRR 191
Query: 183 VSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLHDIARNGS---TVILTIHQPSSRIQ 239
VSI ++I+ P LLFLDEPTSGLDS ++ VI ++ + + T++ +IHQPS+ I
Sbjct: 192 VSICIEILTHPRLLFLDEPTSGLDSAASYHVISRISSLNKKDGIQRTIVASIHQPSNEIF 371
Query: 240 LLLDHLIILARGQLMF 255
L L +L+ G+ ++
Sbjct: 372 QLFHSLCLLSSGKTVY 419
>TC10750 similar to UP|AAP80385 (AAP80385) ABC transporter, partial (38%)
Length = 807
Score = 89.0 bits (219), Expect = 2e-18
Identities = 55/215 (25%), Positives = 104/215 (47%), Gaps = 5/215 (2%)
Frame = +1
Query: 454 SYIGETWILMRRNFTNIRRTPELFLSRLMVLTFMGVMMATMFHNPKNTLQGITNRLSFFI 513
S++ +++ L +R+F N+ R + RL++ + + + T++ N I R S
Sbjct: 139 SFLMQSYTLTKRSFINMSRDFGYYWLRLVIYIVVTICIGTIYLNVGTGYNSILARGSCAS 318
Query: 514 FTVCLFFFSSNDAVPAFIQERFIFIRETSHNAYRASCYTIASLITHMPFLALQALAYAAI 573
F F S P+F+++ +F RE + Y + + +++ ++ PFL L I
Sbjct: 319 FVFGFVTFMSIGGFPSFVEDMKVFQRERLNGHYGVTAFVVSNTVSATPFLVLITFLSGTI 498
Query: 574 VWFALELRGPF---IYFFLVLFISLLSTNSFVVFVSSIVPNYILGYAAVIAFTALFFLFC 630
+F + L F ++F L L+ S+ S ++ ++SIVPN+++G +F L
Sbjct: 499 CYFMVRLHPGFSHYVFFVLCLYASVTVVESLMMAIASIVPNFLMGIIIGAGIQGIFMLVS 678
Query: 631 GYFLSSEDIPL-YWRW-MNKVSTMTYPYEGLLMNE 663
GYF DIP WR+ M+ +S + +G N+
Sbjct: 679 GYFRLPHDIPKPVWRYPMSYISFHFWALQGQYQND 783
>BP055095
Length = 538
Score = 80.5 bits (197), Expect = 9e-16
Identities = 43/140 (30%), Positives = 81/140 (57%), Gaps = 1/140 (0%)
Frame = +3
Query: 154 IEQLGLSSSRNTYIGDEGTRGVSGGERRRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSV 213
++ LGL + IGDE RG+SGG+++RV+ G ++ LF+DE ++GLDS++ +
Sbjct: 39 LKVLGLDICADVMIGDEMRRGISGGQKKRVTTGEMLVGPAKALFMDEISTGLDSSTTFQI 218
Query: 214 IEKLHDIAR-NGSTVILTIHQPSSRIQLLLDHLIILARGQLMFQGSLKDVGHHLNRMGRK 272
+ + + T+++++ QP+ L D +I+L+ GQ+++QG ++V MG K
Sbjct: 219 CKFMRQMVHIMDVTMVISLLQPAPETFELFDDIILLSEGQIVYQGPRENVLEFFEYMGFK 398
Query: 273 IPKGENPIENLIDVIQEYDQ 292
P+ + + L +V + DQ
Sbjct: 399 CPERKGAADFLQEVTSKKDQ 458
>TC9902 similar to UP|BAD07483 (BAD07483) PDR-type ABC transporter 1,
partial (19%)
Length = 1207
Score = 79.0 bits (193), Expect = 3e-15
Identities = 59/291 (20%), Positives = 125/291 (42%), Gaps = 12/291 (4%)
Frame = +1
Query: 441 NSSQEHLGPKFANSYIGETWILMRRNFTNIRRTPELFLSRLMVLTFMGVMMATMFHN--- 497
+S + +F+ ++ + + + + R P R TF+ VM T+F +
Sbjct: 64 DSKDLYFATQFSQPFLIQCQACLWKQRWSYWRNPPYTAVRFFFTTFIAVMFGTIFWDLGG 243
Query: 498 PKNTLQGITNRL-SFFIFTVCLFFFSSNDAVPAFIQERFIFIRETSHNAYRASCYTIASL 556
Q + N + S + + L +S+ P ER +F RE + Y A Y A +
Sbjct: 244 KHKRRQDLLNAVGSMYSAVLFLGVQNSSSVQPVVSVERTVFYREKAAGMYSALPYAFAQI 423
Query: 557 ITHMPFLALQALAYAAIVWFALEL---RGPFIYFFLVLFISLLSTNSFVVFVSSIVPNYI 613
+ +P++ QA+ Y IV+ + F ++ ++ +LL + + ++ PN+
Sbjct: 424 LVELPYIFFQAVTYGVIVYAMIGFDWTAEKFFWYLFFMYFTLLYFTFYGMMGVAVTPNHH 603
Query: 614 LGYAAVIAFTALFFLFCGYFLSSEDIPLYWRWMNKVSTMTYPYEGLLMNEYQTNETFGSN 673
+ AF A++ LF G+ + IP++WRW + + GL+ +++ T
Sbjct: 604 VASIVAAAFYAIWNLFSGFVVPRPSIPVWWRWYYWACPVAWTIYGLIASQFGDITTVMDT 783
Query: 674 DG-----VSITGFDILKSLHIGTEEIKKRNNVLIMLGWAVLYRILFYIILR 719
+G + + + +K IG +++ G A+L+ +F + ++
Sbjct: 784 EGGKTVKMFLEDYYGIKHSFIGV-------CAVVVPGVAILFAFIFAVAIK 915
>BP057421
Length = 547
Score = 71.2 bits (173), Expect = 5e-13
Identities = 47/160 (29%), Positives = 82/160 (50%), Gaps = 32/160 (20%)
Frame = +3
Query: 93 GKVSLDGNSVNASLIKRTSAYIMQEDRLFPMLTVYETLMFAA-----DFRLGPLSAVDKR 147
G+++ +G+ +N + ++T+AYI Q D +TV ETL F+A R LS + +R
Sbjct: 51 GEITYNGHKLNEFVPRKTAAYISQNDVHVGEMTVKETLDFSARCQGVGTRYDXLSELGRR 230
Query: 148 QR---------------------------VEKLIEQLGLSSSRNTYIGDEGTRGVSGGER 180
++ + ++ LGL ++T +GDE RGVSGG++
Sbjct: 231 EKEAGIFPEAELDLFMKATALKGTESSLITDYTLKILGLDICKDTIVGDEMHRGVSGGQK 410
Query: 181 RRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLHDI 220
+RV+ G I+ LF+DE ++GLDS++ +++ L I
Sbjct: 411 KRVTTGEMIVGPTKTLFMDEISTGLDSSTTFQIVKCLQQI 530
>AV424518
Length = 261
Score = 69.7 bits (169), Expect = 2e-12
Identities = 33/85 (38%), Positives = 56/85 (65%), Gaps = 3/85 (3%)
Frame = +1
Query: 525 DAVPAFIQERFIFIRETSHNAYRASCYTIASLITHMPFLALQALAYAAIVWFALELRG-- 582
+A+P F+QER+IF+RET++NAYR S Y ++ + +P L + A+AA ++++ L G
Sbjct: 7 EAIPVFLQERYIFMRETAYNAYRRSSYVLSHSVIALPALIFLSFAFAATTFWSVGLAGGT 186
Query: 583 -PFIYFFLVLFISLLSTNSFVVFVS 606
F ++FL + S + +SFV F+S
Sbjct: 187 SGFFFYFLTIVASFWAGSSFVTFLS 261
>BP029778
Length = 438
Score = 69.3 bits (168), Expect = 2e-12
Identities = 40/113 (35%), Positives = 64/113 (56%), Gaps = 1/113 (0%)
Frame = -2
Query: 27 FESLTYTVTKKKKVDGKWSNED-VDLLHDITGYAPKGCITAVMGPSGAGKSTLLDGLAGR 85
F ++ Y V +++ + E + LL +++G G +TA+MG SGAGK+TL+D LAGR
Sbjct: 353 FHNVNYYVDMPQEIRKQGIIETKLKLLSNVSGVFAPGVLTALMGSSGAGKTTLMDVLAGR 174
Query: 86 IASGSLKGKVSLDGNSVNASLIKRTSAYIMQEDRLFPMLTVYETLMFAADFRL 138
G ++G + + G R + Y+ Q D P +TV E+L+F+A RL
Sbjct: 173 KTGGYIEGDIKISGYPKVQHTFARVAGYVEQNDIHSPQVTVEESLLFSALLRL 15
>TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partial (17%)
Length = 430
Score = 68.9 bits (167), Expect = 3e-12
Identities = 40/100 (40%), Positives = 62/100 (62%), Gaps = 3/100 (3%)
Frame = +1
Query: 51 LLHDITGYAPKGCITAVMGPSGAGKSTLLDGLAGRIASG---SLKGKVSLDGNSVNASLI 107
LL +++G A G + A+MGPSG+GK+TLL+ LAG++A+ L G + +G + +
Sbjct: 106 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLAASPRLHLSGLLEFNGKPGSKNAY 285
Query: 108 KRTSAYIMQEDRLFPMLTVYETLMFAADFRLGPLSAVDKR 147
K AY+ QED F LTV ETL A + +L +S+ ++R
Sbjct: 286 K--FAYVRQEDLFFSQLTVRETLSLATELQLPNISSAEER 399
>TC11137 similar to GB|CAD59563.1|27368811|OSA535041 PDR-like ABC
transporter {Oryza sativa (japonica cultivar-group);},
partial (15%)
Length = 786
Score = 63.5 bits (153), Expect = 1e-10
Identities = 34/118 (28%), Positives = 58/118 (48%), Gaps = 3/118 (2%)
Frame = +1
Query: 551 YTIASLITHMPFLALQALAYAAIVWFALELR---GPFIYFFLVLFISLLSTNSFVVFVSS 607
Y IA + T +P++ Q Y+ IV+ + F ++F V F S L + + S
Sbjct: 268 YAIAQVFTEIPYVFAQTTFYSLIVYAMVSFDWKVDKFFWYFFVSFFSFLYFTYYGMMTVS 447
Query: 608 IVPNYILGYAAVIAFTALFFLFCGYFLSSEDIPLYWRWMNKVSTMTYPYEGLLMNEYQ 665
I PN+ + AF LF LF G+F+ IP +W W + + + GL++++Y+
Sbjct: 448 ITPNHQVASIFAAAFYGLFNLFSGFFIPRPKIPGWWVWYYWICPVAWTVYGLIVSQYR 621
>CB827630
Length = 405
Score = 54.7 bits (130), Expect = 5e-08
Identities = 37/126 (29%), Positives = 63/126 (49%)
Frame = +2
Query: 137 RLGPLSAVDKRQRVEKLIEQLGLSSSRNTYIGDEGTRGVSGGERRRVSIGVDIIHGPSLL 196
RL L + Q VE+ ++ L L + D+ SGG +RR+ + + +I P ++
Sbjct: 32 RLKNLKGLVLTQAVEESLKSLNLFHGG---VADKQAGKYSGGMKRRLIVAISLIGDPRVV 202
Query: 197 FLDEPTSGLDSTSALSVIEKLHDIARNGSTVILTIHQPSSRIQLLLDHLIILARGQLMFQ 256
++DEP++GLD S S + + +A+ +ILT H + L D L I G L
Sbjct: 203 YMDEPSTGLDPASRKS-LWNVVKLAKRDRAIILTTHS-MEEAEALCDRLGIXVNGSLQCV 376
Query: 257 GSLKDV 262
G+ K++
Sbjct: 377 GNAKEL 394
>BP055390
Length = 488
Score = 54.3 bits (129), Expect = 7e-08
Identities = 34/94 (36%), Positives = 56/94 (59%)
Frame = +3
Query: 165 TYIGDEGTRGVSGGERRRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLHDIARNG 224
T +G G + +SGG+++R++I I+ P +L LDE TS LD+ S V E L + A +G
Sbjct: 204 TQVGQRGVQ-LSGGQKQRIAIARAILKNPPILLLDEATSALDTESEKLVQEAL-ETAMHG 377
Query: 225 STVILTIHQPSSRIQLLLDHLIILARGQLMFQGS 258
TVIL H+ S+ + D + ++ GQ++ G+
Sbjct: 378 RTVILIAHRLSTVVN--ADVIAVVENGQVVETGT 473
>TC17528 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;}, partial (20%)
Length = 442
Score = 52.4 bits (124), Expect = 3e-07
Identities = 30/123 (24%), Positives = 62/123 (50%), Gaps = 5/123 (4%)
Frame = +1
Query: 546 YRASCYTIASLITHMPFLALQALAYAAIVWFALELR---GPFIYFFLVLFISLLSTNSFV 602
Y + Y +A+ ++ PFL AL + I + ++ R F++F L ++ S+ S +
Sbjct: 22 YGVAAYILANFLSSFPFLVAIALTTSTITYNMVKFRPGLSHFVFFTLNIYSSISVIESLM 201
Query: 603 VFVSSIVPNYILGYAAVIAFTALFFLFCGYFLSSEDIPL-YWRW-MNKVSTMTYPYEGLL 660
+ V+S+VPN+++G + + G+F D+P WR+ ++ +S + +G
Sbjct: 202 MVVASLVPNFLMGIITGAGIIGIMMMTSGFFRLLSDLPKPVWRYPISYISYGAWAIQGSY 381
Query: 661 MNE 663
N+
Sbjct: 382 KND 390
>TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proetin, partial
(9%)
Length = 712
Score = 49.7 bits (117), Expect = 2e-06
Identities = 28/66 (42%), Positives = 43/66 (64%)
Frame = +2
Query: 164 NTYIGDEGTRGVSGGERRRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLHDIARN 223
+T +G+ GT +SGG+++RV+I II P++L LDE TS LD+ S V + L + N
Sbjct: 77 DTVVGERGTL-LSGGQKQRVAIARAIIKTPNILLLDEATSALDAESERVVQDALDKVMVN 253
Query: 224 GSTVIL 229
+TVI+
Sbjct: 254 RTTVIV 271
>BG662257
Length = 336
Score = 49.7 bits (117), Expect = 2e-06
Identities = 29/78 (37%), Positives = 45/78 (57%)
Frame = +1
Query: 159 LSSSRNTYIGDEGTRGVSGGERRRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLH 218
L +T +GD G + +SGG+++RV+I I+ P +L LDE TS LD+ S V++
Sbjct: 94 LQQGYDTVVGDRGIQ-LSGGQKQRVAIARAIVKSPKILLLDEATSALDAESE-KVVQDAL 267
Query: 219 DIARNGSTVILTIHQPSS 236
D R T I+ H+ S+
Sbjct: 268 DRVRVDRTTIVVAHRLST 321
>AV776921
Length = 435
Score = 48.5 bits (114), Expect = 4e-06
Identities = 41/143 (28%), Positives = 71/143 (48%), Gaps = 4/143 (2%)
Frame = +2
Query: 45 SNEDVDLLHDITGYAPKGCITAVMGPSGAGKSTLLDGLAGRIASGSLKGKVSLDGNSV-- 102
S DV++L+ + P G I A++G SG+GKST++ L R L G + LDGN +
Sbjct: 23 SRPDVEILNKLCLDIPSGKIVALVGGSGSGKSTVI-SLIERFYE-PLSGDILLDGNDIRD 196
Query: 103 -NASLIKRTSAYIMQEDRLFPMLTVYETLMFAAD-FRLGPLSAVDKRQRVEKLIEQLGLS 160
+ +++ + QE LF ++ E +++ D L L K + I L
Sbjct: 197 LDLKWLRQQIGLVNQEPALF-ATSIKENILYGKDNATLEELKRAVKLSDAQSFIN--NLP 367
Query: 161 SSRNTYIGDEGTRGVSGGERRRV 183
T +G+ G + +SGG+++R+
Sbjct: 368 ERLETQVGERGIQ-LSGGQKQRI 433
>AV422962
Length = 382
Score = 47.0 bits (110), Expect = 1e-05
Identities = 28/117 (23%), Positives = 58/117 (48%), Gaps = 5/117 (4%)
Frame = +3
Query: 554 ASLITHMPFLALQALAYAAIVWFALELRGPFIYFFLVLFISLLSTNSFV-----VFVSSI 608
+ L+ P A L + A+++ L + F F +++ SF + V ++
Sbjct: 3 SKLLAEAPIGAAFPLMFGAVLYPMARLHPTLMRFGK--FCGIVTMESFAASAMGLTVGAM 176
Query: 609 VPNYILGYAAVIAFTALFFLFCGYFLSSEDIPLYWRWMNKVSTMTYPYEGLLMNEYQ 665
VP A + +F +F GY+++ E+ P+ +RW+ VS + + ++GL +NE++
Sbjct: 177 VPTTEAAMAVGPSLMTVFIVFGGYYVNRENTPIIFRWIPSVSLIRWAFQGLCVNEFR 347
>TC18135 homologue to UP|Q9MAY4 (Q9MAY4) ABC transporter homolog, partial
(50%)
Length = 968
Score = 46.2 bits (108), Expect = 2e-05
Identities = 42/166 (25%), Positives = 69/166 (41%), Gaps = 10/166 (6%)
Frame = +3
Query: 67 VMGPSGAGKSTLLDGLAGRIASGSLKGKVSLDGNSVNASLIKRTSAYIMQEDRLFPMLTV 126
++G +G GKSTLL + R + + AS + A + ++ +
Sbjct: 390 LLGLNGCGKSTLLTAIGHRELPIPEHMDIYHLTREIEASDMSALEAVVSCDEERLRLEKE 569
Query: 127 YETLMFAADF----------RLGPLSAVDKRQRVEKLIEQLGLSSSRNTYIGDEGTRGVS 176
ETL D RL L A +R +++ LG + TR S
Sbjct: 570 AETLAAQDDGGGETLERIYERLEALDASTAEKRAAEILHGLGFDKKMQA----KKTRDFS 737
Query: 177 GGERRRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLHDIAR 222
GG R R+++ + P++L LDEPT+ LD + + + E L + R
Sbjct: 738 GGWRMRIALARALFINPTILLLDEPTNHLDLEACVWLEESLKNFDR 875
>TC15722 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transporter,
partial (37%)
Length = 624
Score = 45.4 bits (106), Expect = 3e-05
Identities = 21/64 (32%), Positives = 39/64 (60%)
Frame = +3
Query: 169 DEGTRGVSGGERRRVSIGVDIIHGPSLLFLDEPTSGLDSTSALSVIEKLHDIARNGSTVI 228
D+ + +SGG +RR+++ + ++ P LL LDEP +GLD + V++ L + + TV+
Sbjct: 51 DKNPQSLSGGYKRRLALAIQLVQVPDLLILDEPLAGLDWKARADVVKLLKHLKKE-LTVL 227
Query: 229 LTIH 232
+ H
Sbjct: 228 VVSH 239
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,058,960
Number of Sequences: 28460
Number of extensions: 176057
Number of successful extensions: 1271
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 1248
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1263
length of query: 727
length of database: 4,897,600
effective HSP length: 97
effective length of query: 630
effective length of database: 2,136,980
effective search space: 1346297400
effective search space used: 1346297400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC126010.8


