

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.4 - phase: 0
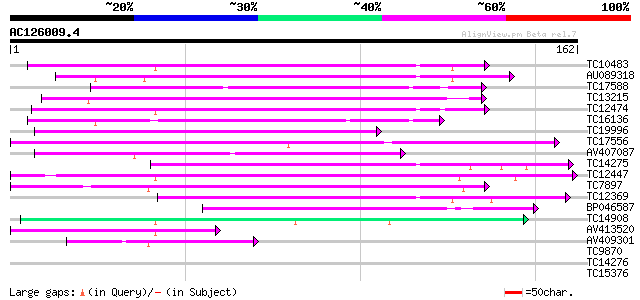
(162 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10483 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (... 96 4e-21
AU089318 92 4e-20
TC17588 similar to UP|BCP_PEA (Q41001) Blue copper protein precu... 89 5e-19
TC13215 similar to UP|Q39040 (Q39040) Lamin, partial (15%) 86 4e-18
TC12474 weakly similar to UP|O82576 (O82576) Blue copper protein... 84 9e-18
TC16136 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein,... 81 1e-16
TC19996 weakly similar to UP|ENL1_ARATH (Q9SK27) Early nodulin-l... 78 8e-16
TC17556 similar to UP|Q39131 (Q39131) Lamin (AT5G15350/F8M21_240... 73 3e-14
AV407087 72 4e-14
TC14275 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (... 69 5e-13
TC12447 weakly similar to GB|AAM63773.1|21555085|AY086719 phytoc... 57 2e-09
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 55 5e-09
TC12369 weakly similar to UP|ENL1_ARATH (Q9SK27) Early nodulin-l... 53 3e-08
BP046587 52 4e-08
TC14908 weakly similar to PIR|A86218|A86218 protein T27G7.18 [im... 52 7e-08
AV413520 49 3e-07
AV409301 39 6e-04
TC9870 weakly similar to UP|NO16_MEDTR (P93328) Early nodulin 16... 35 0.005
TC14276 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (... 34 0.014
TC15376 weakly similar to UP|Q7SYH2 (Q7SYH2) Cystatin domain fet... 32 0.070
>TC10483 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (24%)
Length = 552
Score = 95.5 bits (236), Expect = 4e-21
Identities = 54/140 (38%), Positives = 74/140 (52%), Gaps = 8/140 (5%)
Frame = +3
Query: 6 VVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFD----YTQWAQDKVFRVGDNLVFNYD 61
V+L+ ++ L + A T ++VGD GWT+ YT WA +K F VGD LVFN+
Sbjct: 105 VLLLALLAATTLFHTSSAQTRHVVGDSTGWTIPTGGASFYTNWAANKTFSVGDTLVFNFA 284
Query: 62 PSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVI 121
+HNV KV + F SC+ +STG + L G + ++C HCSA Q KL +
Sbjct: 285 NGQHNVAKVTKSAFDSCSGSSPVTLISTGPATVTLTETGVQHFICAFGGHCSAGQ-KLAV 461
Query: 122 TVLA----EGAPAPSPPPSS 137
T +A APAP P P S
Sbjct: 462 TXVAGKASNNAPAPVPAPKS 521
>AU089318
Length = 641
Score = 92.0 bits (227), Expect = 4e-20
Identities = 49/139 (35%), Positives = 78/139 (55%), Gaps = 8/139 (5%)
Frame = +3
Query: 14 MVLLSSVAIA-ATDYIVGDDKGWTV--DFDYTQWAQDKVFRVGDNLVFNYDPSRHNVFKV 70
MV++++V ++ A + VGD GWT+ + DY +WA F+VGD ++F Y+ HNV +V
Sbjct: 66 MVVMAAVQVSYAAVHKVGDSAGWTILGNVDYKKWAAPXNFQVGDTIIFEYNAQFHNVMRV 245
Query: 71 NGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLA----- 125
++ +C STG D I++ G ++ CGV HC A Q K+ I V++
Sbjct: 246 THAMYXTCNASSPIATFSTGNDSIKITNHGHHFFFCGVPGHCQAGQ-KVDINVISVSAAT 422
Query: 126 EGAPAPSPPPSSDAHSVVS 144
AP P+P PSS + S+ +
Sbjct: 423 AAAPTPTPAPSSPSSSLAT 479
>TC17588 similar to UP|BCP_PEA (Q41001) Blue copper protein precursor,
partial (61%)
Length = 747
Score = 88.6 bits (218), Expect = 5e-19
Identities = 50/113 (44%), Positives = 65/113 (57%)
Frame = +1
Query: 24 ATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCTFPPK 83
AT Y VGD GW DY+ W DK F VGD+LVFNY + H V +V + ++SCT
Sbjct: 4 ATVYSVGDTSGWAAGADYSTWTSDKTFAVGDSLVFNYG-AGHTVDEVKESDYKSCTTGNS 180
Query: 84 NEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLAEGAPAPSPPPS 136
S+GK I LKT G +++C V HCS MKL +TV +GA + + P S
Sbjct: 181 LSTDSSGKTTIALKTAGTHYFICSVPGHCSG-GMKLAVTV--KGAASFTTPSS 330
>TC13215 similar to UP|Q39040 (Q39040) Lamin, partial (15%)
Length = 452
Score = 85.5 bits (210), Expect = 4e-18
Identities = 46/132 (34%), Positives = 75/132 (55%), Gaps = 5/132 (3%)
Frame = +2
Query: 10 LSISMVLLSSVA-----IAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDPSR 64
L+ S++LL+ + + ATD+IVG ++GW +YT WA ++ F VGD + F Y ++
Sbjct: 74 LTCSLLLLTFITFTISPVTATDHIVGANRGWNPGQNYTLWANNQTFYVGDFISFRYQKNQ 253
Query: 65 HNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVL 124
+NVF+VN T + +CT S+GKD I L GR +++CG + ++ +V+ L
Sbjct: 254 YNVFEVNQTGYDNCTTEGAFGNYSSGKDFIMLNKTGRHYFICGNGQCFNGMKVSVVVHPL 433
Query: 125 AEGAPAPSPPPS 136
A +PP S
Sbjct: 434 A------APPTS 451
>TC12474 weakly similar to UP|O82576 (O82576) Blue copper protein
(Fragment), partial (44%)
Length = 818
Score = 84.3 bits (207), Expect = 9e-18
Identities = 50/134 (37%), Positives = 69/134 (51%), Gaps = 3/134 (2%)
Frame = +1
Query: 7 VLILSISMVLLSSVAIAATDYIVGDDKGWTVDFD---YTQWAQDKVFRVGDNLVFNYDPS 63
+L+ ++ L + A T ++VGD GW V + YT WA F VGD LVFNY S
Sbjct: 97 LLLALLAATALFHCSSAQTRHVVGDSAGWFVPGNTSFYTSWAAKNTFAVGDTLVFNYAAS 276
Query: 64 RHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITV 123
HNV +V T + SC +T + +K G +++CGV HC Q KL I V
Sbjct: 277 AHNVEEVTKTNYDSCNSTSPIATYTTPPVTVTIKKTGAHYFICGVPGHCLGDQ-KLSINV 453
Query: 124 LAEGAPAPSPPPSS 137
A G+ A +PP S+
Sbjct: 454 -ASGSSAATPPSST 492
>TC16136 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein, partial
(82%)
Length = 872
Score = 80.9 bits (198), Expect = 1e-16
Identities = 46/122 (37%), Positives = 70/122 (56%), Gaps = 3/122 (2%)
Frame = +1
Query: 6 VVLILSISMVLLSSVAIA---ATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDP 62
VV+ + IS++ L ++ + A Y VG GW+ + D W K FR GD L+FNYD
Sbjct: 106 VVVTVGISLLCLLALQVEHANAATYTVGGPAGWSFNTD--TWPNGKKFRAGDVLIFNYDS 279
Query: 63 SRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVIT 122
+ HNV V+ + ++SCT P + LS+GKD I+L G+ +++C HC + MK+ I
Sbjct: 280 TTHNVVAVDQSGYKSCTTPAGAKVLSSGKDQIRL-GRGQNYFICNCPGHCQS-GMKVAIN 453
Query: 123 VL 124
L
Sbjct: 454 AL 459
>TC19996 weakly similar to UP|ENL1_ARATH (Q9SK27) Early nodulin-like protein
1 precursor (Phytocyanin-like protein), partial (36%)
Length = 458
Score = 77.8 bits (190), Expect = 8e-16
Identities = 34/99 (34%), Positives = 56/99 (56%)
Frame = +3
Query: 8 LILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDPSRHNV 67
++ S+ ++L++ + D+ VGD GW + DYT WA K F+VGDNLVF Y S H V
Sbjct: 87 MVASLLVLLVAFPTVFGADHTVGDASGWNIGVDYTTWASGKTFKVGDNLVFTYSSSLHGV 266
Query: 68 FKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVC 106
+V+ + ++SC+ + S G + L G +++C
Sbjct: 267 DEVDESSYKSCSTSSPIKTYSDGNTKVALTKAGTLYFIC 383
>TC17556 similar to UP|Q39131 (Q39131) Lamin (AT5G15350/F8M21_240), partial
(68%)
Length = 595
Score = 72.8 bits (177), Expect = 3e-14
Identities = 48/159 (30%), Positives = 80/159 (50%), Gaps = 2/159 (1%)
Frame = +2
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNY 60
+ S +VL+ VLL ++A ++VGD K W + +YT WA+DK F + D L F Y
Sbjct: 98 LGSPAMVLLFLGFAVLLMVPEVSAKRWLVGDGKFWNPNVNYTVWARDKHFYIDDWLFFVY 277
Query: 61 DPSRHNVFKVNGTLFQSC--TFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMK 118
D ++ NV +VN T +++C P N G+D++ L +++ G + MK
Sbjct: 278 DRNQMNVLEVNKTNYENCIAEHPIHNWTTGAGRDVVPLNVTRHYYFISG--NGFCYGGMK 451
Query: 119 LVITVLAEGAPAPSPPPSSDAHSVVSSLFGVVMAIMVAI 157
L + V P + P + A S+ S ++M ++ AI
Sbjct: 452 LAVRVEKLPPPPKAAPEKAGAPSLSSRGAILLMPVVFAI 568
>AV407087
Length = 363
Score = 72.4 bits (176), Expect = 4e-14
Identities = 36/107 (33%), Positives = 56/107 (51%), Gaps = 1/107 (0%)
Frame = +3
Query: 8 LILSISMVLLSSVAIAATDYIVGDDKG-WTVDFDYTQWAQDKVFRVGDNLVFNYDPSRHN 66
+I+ +S+V ATD+IVG G W + W + F VGDNL+F Y P+ HN
Sbjct: 39 VIIRLSLVATLIKLAMATDHIVGGPNGGWDATSNLQSWGTSQQFSVGDNLIFQYQPN-HN 215
Query: 67 VFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCS 113
V +V + SC ++ S G I L + G+++++CG HC+
Sbjct: 216 VIEVTKADYDSCQATSPIQSYSDGATTIPLSSPGKRYFICGAIGHCT 356
>TC14275 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (25%)
Length = 562
Score = 68.6 bits (166), Expect = 5e-13
Identities = 47/134 (35%), Positives = 61/134 (45%), Gaps = 13/134 (9%)
Frame = +1
Query: 41 YTQWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEG 100
Y WA K F+VGD LVFNY S HNV +V + SC +T + +K G
Sbjct: 4 YRTWASGKTFKVGDILVFNYAASAHNVEEVTKQKYDSCNSTSPIATYTTPPVRVTIKKTG 183
Query: 101 RKWYVCGVADHCSARQMKLVITVLAEGAPA-------PSPPPSSDA----HSVVSSL--F 147
+++CGV HC Q KL I V + A PSP PSS S SL
Sbjct: 184 AHYFICGVPGHCLGGQ-KLSINVTGGSSTATPPSSASPSPSPSSSTTPPQDSAAGSLGAA 360
Query: 148 GVVMAIMVAIAVIF 161
GV + ++ I +F
Sbjct: 361 GVFASTVMTITAVF 402
>TC12447 weakly similar to GB|AAM63773.1|21555085|AY086719
phytocyanin-related protein-like {Arabidopsis thaliana;}
, partial (56%)
Length = 735
Score = 56.6 bits (135), Expect = 2e-09
Identities = 46/173 (26%), Positives = 73/173 (41%), Gaps = 11/173 (6%)
Frame = +3
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFD----YTQWAQDKVFRVGDNL 56
MA S L+L + LL + A + G W V QWA+ F+VGD L
Sbjct: 51 MAGSSASLLL---LFLLFGFSAAKELLVGGKTDAWKVPSSEADSLNQWAEKSRFKVGDYL 221
Query: 57 VFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQ 116
V+ YD + +V +VN + SC E G ++L G +++ G HC Q
Sbjct: 222 VWKYDGGKDSVLQVNKEDYGSCNTSNPIEEYKDGNTKVKLDRPGPHYFISGAKGHCEKGQ 401
Query: 117 MKLVITVLAEG------APAPSPPPSSDAHSVV-SSLFGVVMAIMVAIAVIFA 162
V+ + + +PA SP + +V +S V+ + VA+ + A
Sbjct: 402 KLFVVVMTPKHSRDRAISPASSPAELEEGPAVAPTSSATVLQSGFVAVLGVLA 560
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 55.5 bits (132), Expect = 5e-09
Identities = 38/140 (27%), Positives = 58/140 (41%), Gaps = 3/140 (2%)
Frame = +3
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVD--FDYTQWAQDKVFRVGDNLVF 58
M R + +L + LL S A + VG KGW + YT WA F++ D +VF
Sbjct: 141 MEFQRPLCLLFLLFSLLPSSQ--AHKFYVGGSKGWIPNPSESYTLWAGRNRFQIKDTIVF 314
Query: 59 NYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMK 118
Y +V +V F +C + G G +++ G +C Q
Sbjct: 315 KYTKGSDSVLEVKKEDFDNCNKANPIKKFEDGDTEFTFDRSGPFYFISGKDGNCEKGQKM 494
Query: 119 LVITVLAEGA-PAPSPPPSS 137
+++ + G P PSPPP S
Sbjct: 495 ILVVISPRGTQPPPSPPPKS 554
>TC12369 weakly similar to UP|ENL1_ARATH (Q9SK27) Early nodulin-like protein
1 precursor (Phytocyanin-like protein), partial (47%)
Length = 574
Score = 52.8 bits (125), Expect = 3e-08
Identities = 36/134 (26%), Positives = 65/134 (47%), Gaps = 16/134 (11%)
Frame = +1
Query: 43 QWAQDKVFRVGDNLVFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRK 102
+WAQ F++GD+LV+ YD + +V +V+ ++ C + + G ++L G
Sbjct: 16 KWAQRDRFQIGDHLVWKYDGGKDSVLEVSKEDYEKCNISSPIKEYNDGNTKVKLDHPGPF 195
Query: 103 WYVCGVADHCSARQMKLVITVLA-----------EGAPAPSPPPS-----SDAHSVVSSL 146
+++ G HC Q KLV+ VL+ APA +P P+ + A S VSS
Sbjct: 196 YFISGAKGHCEKGQ-KLVVVVLSPRGGRNTGTDVSPAPASAPVPTGFEGPAVAPSPVSSA 372
Query: 147 FGVVMAIMVAIAVI 160
+ +++ + V+
Sbjct: 373 TALQGGLVMLMGVL 414
>BP046587
Length = 502
Score = 52.4 bits (124), Expect = 4e-08
Identities = 32/96 (33%), Positives = 50/96 (51%)
Frame = -3
Query: 56 LVFNYDPSRHNVFKVNGTLFQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSAR 115
+ F Y +++NVF+VN T + +CT S+GKD I L GR +++CG +
Sbjct: 464 VAFRYQKNQYNVFEVNQTGYDNCTTEGAFGNYSSGKDFIMLNKTGRHYFICGNGQCFNGM 285
Query: 116 QMKLVITVLAEGAPAPSPPPSSDAHSVVSSLFGVVM 151
++ +V+ LA AP P S+ HS S VV+
Sbjct: 284 KVSVVVHPLA--AP---PTSSTGEHSTPKSSAPVVL 192
>TC14908 weakly similar to PIR|A86218|A86218 protein T27G7.18 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(68%)
Length = 1219
Score = 51.6 bits (122), Expect = 7e-08
Identities = 41/166 (24%), Positives = 63/166 (37%), Gaps = 21/166 (12%)
Frame = +2
Query: 4 SRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFD-----YTQWAQDKVFRVGDNLVF 58
S+V L + + S A +Y VGD GW + + Y +W +K F +GD L+F
Sbjct: 134 SKVSCFLLLLIFCFSGSVEAYKNYTVGDSLGWFDNLEKPHVNYQKWVANKEFSLGDFLIF 313
Query: 59 NYDPSRHNVFKVNGTLFQSCTF--------------PPKNEALSTGKDIIQLKTEGRKWY 104
N D + V N T ++ C + P N + + L EG ++
Sbjct: 314 NTDTNHTVVQTYNFTTYKQCDYNDAQDKDTTQWSSSDPSNTEIHPVTAAVPLVKEGMNYF 493
Query: 105 VCG--VADHCSARQMKLVITVLAEGAPAPSPPPSSDAHSVVSSLFG 148
D C Q + +G P PS D+ S S + G
Sbjct: 494 FSSDYDGDQCKNGQHFKINVTYGQGLPKSLRSPSEDSPSPASPVSG 631
>AV413520
Length = 280
Score = 49.3 bits (116), Expect = 3e-07
Identities = 26/65 (40%), Positives = 38/65 (58%), Gaps = 5/65 (7%)
Frame = +1
Query: 1 MASSRVVLILSISMVLLSSVAIAATDYIVGDDKGWTVDFD-----YTQWAQDKVFRVGDN 55
+ S++ V L +LL AA +++VG KGW+V D + QWA+ F+VGD+
Sbjct: 76 LGSNKTVHALGWFCLLLMVHKSAAYEFVVGGQKGWSVPSDPNFNPFNQWAEKSRFQVGDS 255
Query: 56 LVFNY 60
LVFNY
Sbjct: 256 LVFNY 270
>AV409301
Length = 360
Score = 38.5 bits (88), Expect = 6e-04
Identities = 24/57 (42%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Frame = +2
Query: 17 LSSVAIAATDYIVGDDKGWTVD--FDYTQWAQDKVFRVGDNLVFNYDPSRHNVFKVN 71
L S + A T Y+ G D GW ++ +Y QWA F+V DNLVF Y +V VN
Sbjct: 176 LFSGSQAYTFYVGGKD-GWVLNPSENYNQWAGRNRFQVNDNLVFKYKTGSDSVLVVN 343
>TC9870 weakly similar to UP|NO16_MEDTR (P93328) Early nodulin 16 precursor
(N-16), partial (31%)
Length = 581
Score = 35.4 bits (80), Expect = 0.005
Identities = 30/134 (22%), Positives = 53/134 (39%), Gaps = 28/134 (20%)
Frame = +1
Query: 56 LVFNYDPSRHNVFKVNGTLFQSCTFPPKN-EALSTGKDIIQLKTEGRKWYVCGVADHCSA 114
L+F YD +V VN + C ++ E G + + G ++ G DHC
Sbjct: 1 LIFTYDNETESVHVVNEEDYLKCKVEGEDHEVYLEGYNKVVFNRSGSHLFISGKDDHCKM 180
Query: 115 RQMKLVITVLAE---------------------------GAPAPSPPPSSDAHSVVSSLF 147
+KL + V++ +P PSP P+S+ + +S
Sbjct: 181 G-LKLAVVVMSHRHHTEILSSNSDPSSLSPSPSPAPSSSSSPTPSPSPASNGVASISG-S 354
Query: 148 GVVMAIMVAIAVIF 161
G +M + VA+ ++F
Sbjct: 355 GFIMWMGVALVMLF 396
>TC14276 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (16%)
Length = 624
Score = 33.9 bits (76), Expect = 0.014
Identities = 24/78 (30%), Positives = 32/78 (40%), Gaps = 7/78 (8%)
Frame = +3
Query: 75 FQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLAEGAPA---- 130
+ SC +T + +K G +++CGV HC Q KL I V + A
Sbjct: 18 YDSCNSTSPIATYTTPPVRVTIKKTGAHYFICGVPGHCLGGQ-KLSINVTGGSSTATPPS 194
Query: 131 ---PSPPPSSDAHSVVSS 145
PSP PSS SS
Sbjct: 195 SASPSPSPSSSTTPSPSS 248
>TC15376 weakly similar to UP|Q7SYH2 (Q7SYH2) Cystatin domain fetuin-like
protein, partial (6%)
Length = 582
Score = 31.6 bits (70), Expect = 0.070
Identities = 30/129 (23%), Positives = 57/129 (43%), Gaps = 1/129 (0%)
Frame = +3
Query: 8 LILSISMVLLSSVAIAATDYIVGDDKGWTVDFDYTQWAQDKVFRVGDNLVFNYDPSRHNV 67
L L +++V LSS+ + I DD G + Y W Q L+ + + +
Sbjct: 36 LFLLLALVTLSSITYPS---IAADDGGGGLVSVY--WGQAS--NEEGTLLEACETGNYGI 194
Query: 68 FKVNGTL-FQSCTFPPKNEALSTGKDIIQLKTEGRKWYVCGVADHCSARQMKLVITVLAE 126
+ G + + S P N G D L + +K ++C + +K+++++LA+
Sbjct: 195 VIIEGLIVYNSGNTPTLNLRNHCGSDQQHLCSALQK-----DIEYCQQKNVKVILSILAD 359
Query: 127 GAPAPSPPP 135
+ P+PPP
Sbjct: 360 DSVTPAPPP 386
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,069,439
Number of Sequences: 28460
Number of extensions: 48418
Number of successful extensions: 591
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 550
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 580
length of query: 162
length of database: 4,897,600
effective HSP length: 83
effective length of query: 79
effective length of database: 2,535,420
effective search space: 200298180
effective search space used: 200298180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC126009.4


