

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.1 - phase: 0
(326 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
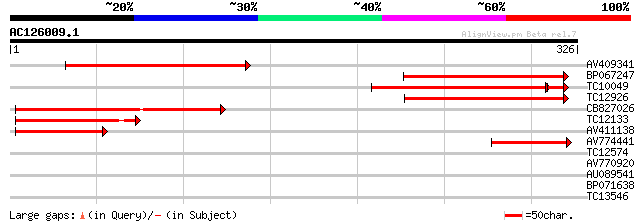
Score E
Sequences producing significant alignments: (bits) Value
AV409341 169 6e-43
BP067247 162 5e-41
TC10049 149 1e-38
TC12926 143 4e-35
CB827026 135 9e-33
TC12133 71 3e-13
AV411138 62 1e-10
AV774441 53 8e-08
TC12574 30 0.42
AV770920 30 0.55
AU089541 29 1.2
BP071638 27 3.6
TC13546 27 4.6
>AV409341
Length = 321
Score = 169 bits (428), Expect = 6e-43
Identities = 80/106 (75%), Positives = 88/106 (82%)
Frame = +2
Query: 33 VFIPIGVASLFASEQVVEVPLRYDDQCLPSLYKDDAMTYIKGNRISKTCTKKLTVKSKMK 92
+FIPIG+ASLFASE+VVEVP RYDDQCLP Y+DDA+ YIK + SKTC KKLTVK+KMK
Sbjct: 2 IFIPIGLASLFASERVVEVPFRYDDQCLPPAYRDDAVAYIKDDSSSKTCIKKLTVKNKMK 181
Query: 93 APIYVYYQLSNFYQNHRHYVKSRDHKQLRSKADENDVGKCFPEDYT 138
AP+YVYYQL NFYQNHR YVKSR QLRSKA E DV C PEDYT
Sbjct: 182 APVYVYYQLDNFYQNHRRYVKSRSDTQLRSKAAEYDVSSCSPEDYT 319
>BP067247
Length = 515
Score = 162 bits (411), Expect = 5e-41
Identities = 73/95 (76%), Positives = 87/95 (90%)
Frame = -3
Query: 227 LPTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYI 286
LPTFRKLYGKIE DLE NDEIT+VIEN+YNTY+FGG KS++LSTTTWIGG+N FLGIAYI
Sbjct: 513 LPTFRKLYGKIENDLEVNDEITMVIENDYNTYEFGGKKSLVLSTTTWIGGRNHFLGIAYI 334
Query: 287 LIGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKN 321
IGG+SL++++ LLMY+MKPRPLGDP YL+WN+N
Sbjct: 333 FIGGMSLLFAIACLLMYVMKPRPLGDPTYLSWNRN 229
>TC10049
Length = 643
Score = 149 bits (375), Expect(2) = 1e-38
Identities = 67/103 (65%), Positives = 84/103 (81%)
Frame = +2
Query: 209 SIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVIL 268
+I LSEQEDLIVWMRTAALPTFRKLYGKIEVDL+ + I+V ++NNYNTY F G K ++L
Sbjct: 14 AIALSEQEDLIVWMRTAALPTFRKLYGKIEVDLDEGENISVKLQNNYNTYSFNGKKKLVL 193
Query: 269 STTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYLMKPRPLG 311
STT+W+GGKNDFLG+AY+ +GGL +L F ++Y +KPR G
Sbjct: 194 STTSWLGGKNDFLGLAYLTVGGLCFFLALAFTIVYFVKPRHTG 322
Score = 27.3 bits (59), Expect(2) = 1e-38
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +3
Query: 310 LGDPRYLTWNKN 321
LGDP YL+WN+N
Sbjct: 318 LGDPSYLSWNRN 353
>TC12926
Length = 580
Score = 143 bits (360), Expect = 4e-35
Identities = 62/94 (65%), Positives = 78/94 (82%)
Frame = +1
Query: 228 PTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYIL 287
PTFRKLYGKIE+DLE D I VV++NNY+TY F G K ++LSTT+W+GGKNDFLGIAY+
Sbjct: 1 PTFRKLYGKIEMDLEKGDVIKVVLQNNYHTYSFNGKKKLVLSTTSWLGGKNDFLGIAYLT 180
Query: 288 IGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKN 321
+GGLS S+VF ++Y +KPR LGDP YL+WN+N
Sbjct: 181 VGGLSFFLSMVFTIVYFVKPRQLGDPSYLSWNRN 282
>CB827026
Length = 542
Score = 135 bits (340), Expect = 9e-33
Identities = 67/121 (55%), Positives = 88/121 (72%)
Frame = +3
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTP VI+ F + +VFIPIGVASL AS VVE+ RY+ C+P
Sbjct: 180 SKFTQQELPACKPILTPRAVISAFLIVTIVFIPIGVASLIASRDVVEIIHRYEADCVPGN 359
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+ D + YI + KTCT+++ V+ +MK+PIYVYYQL NFYQNHR YVKSR+ +QLR
Sbjct: 360 WSSDKVGYIHSS-ADKTCTREIHVEKRMKSPIYVYYQLDNFYQNHRRYVKSRNDEQLRDS 536
Query: 124 A 124
+
Sbjct: 537 S 539
>TC12133
Length = 475
Score = 70.9 bits (172), Expect = 3e-13
Identities = 36/72 (50%), Positives = 51/72 (70%)
Frame = +2
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTP VI+ F + +VF+PIGVASL AS +VVE+ RY+ CL +
Sbjct: 257 SKFTQQELPACKPILTPRAVISAFLLVSVVFVPIGVASLIASRKVVEIVHRYESSCLKGV 436
Query: 64 YKDDAMTYIKGN 75
D+ + YI+ +
Sbjct: 437 --DNKIAYIQSS 466
>AV411138
Length = 428
Score = 62.4 bits (150), Expect = 1e-10
Identities = 32/53 (60%), Positives = 40/53 (75%)
Frame = +1
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYD 56
S+F+QQEL A +PILTP VI+ F + +VFIPIGVASL AS VVE+ RY+
Sbjct: 265 SKFTQQELPACKPILTPRAVISAFLIVTIVFIPIGVASLIASRDVVEIIHRYE 423
>AV774441
Length = 433
Score = 52.8 bits (125), Expect = 8e-08
Identities = 22/46 (47%), Positives = 30/46 (64%)
Frame = -1
Query: 278 NDFLGIAYILIGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKNSK 323
NDFL +AY+ +GGL +L F L+Y ++PRPLGDP Y S+
Sbjct: 433 NDFLALAYLTLGGLCFFLALAFTLLYFVQPRPLGDPSYFVME*ESR 296
>TC12574
Length = 325
Score = 30.4 bits (67), Expect = 0.42
Identities = 11/17 (64%), Positives = 14/17 (81%)
Frame = +2
Query: 252 ENNYNTYQFGGTKSVIL 268
ENN+NTY FGG K ++L
Sbjct: 269 ENNFNTYSFGGKKKLLL 319
>AV770920
Length = 426
Score = 30.0 bits (66), Expect = 0.55
Identities = 10/13 (76%), Positives = 13/13 (99%)
Frame = -1
Query: 272 TWIGGKNDFLGIA 284
+W+GGKNDFLG+A
Sbjct: 426 SWLGGKNDFLGVA 388
>AU089541
Length = 628
Score = 28.9 bits (63), Expect = 1.2
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +1
Query: 23 VIAIFTFIGLVFIPIGVASLFAS 45
VIA F +G +FIP+G+ +L AS
Sbjct: 556 VIATFLLMGFIFIPVGLVTLRAS 624
>BP071638
Length = 406
Score = 27.3 bits (59), Expect = 3.6
Identities = 12/33 (36%), Positives = 20/33 (60%)
Frame = -2
Query: 273 WIGGKNDFLGIAYILIGGLSLVYSLVFLLMYLM 305
+ G K+DFLG+ +I +V++LVF + M
Sbjct: 396 YFGFKHDFLGVVAAVIVAFPVVFALVFAISIKM 298
>TC13546
Length = 415
Score = 26.9 bits (58), Expect = 4.6
Identities = 11/24 (45%), Positives = 17/24 (70%)
Frame = +1
Query: 297 LVFLLMYLMKPRPLGDPRYLTWNK 320
L+ L +++ R LGDP YL+WN+
Sbjct: 88 LICLCLFI---RQLGDPSYLSWNR 150
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,058,065
Number of Sequences: 28460
Number of extensions: 83178
Number of successful extensions: 392
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 391
length of query: 326
length of database: 4,897,600
effective HSP length: 91
effective length of query: 235
effective length of database: 2,307,740
effective search space: 542318900
effective search space used: 542318900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC126009.1


