

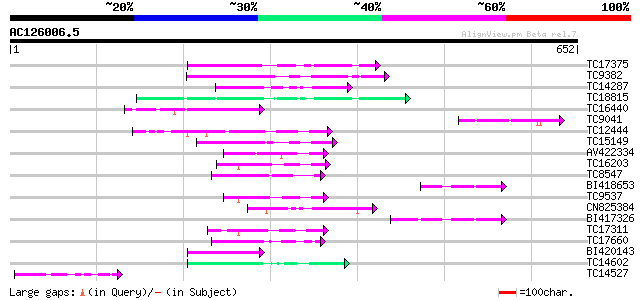
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.5 + phase: 0
(652 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 66 2e-11
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 65 3e-11
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 64 1e-10
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 63 1e-10
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 54 6e-08
TC9041 similar to UP|Q8LLW2 (Q8LLW2) Receptor-like kinase Xa21-b... 52 2e-07
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 52 4e-07
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 49 3e-06
AV422334 47 1e-05
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 46 2e-05
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 46 2e-05
BI418653 46 2e-05
TC9537 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 45 3e-05
CN825384 45 4e-05
BI417326 45 4e-05
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 45 5e-05
TC17660 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, part... 45 5e-05
BI420143 45 5e-05
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 45 5e-05
TC14527 similar to PIR|F86340|F86340 protein F2D10.34 [imported]... 44 6e-05
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 66.2 bits (160), Expect = 2e-11
Identities = 54/226 (23%), Positives = 95/226 (41%), Gaps = 4/226 (1%)
Frame = +3
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ ++G+ +E L + ++ G ++L+ G + Y I G+ LH+
Sbjct: 114 NIVQYYGSELGEESLSVYLEYVSGGSIHKLLQEYGAFKEPVIQNYTRQIVSGLAYLHSRN 293
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIML--SP 322
V +K +N+L+D NG ++D+G+ S I+S + SP
Sbjct: 294 TVHRDIKGANILVDPNGEIKLADFGM-----------------SKHINSAASMLSFKGSP 422
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
++ APE + +G G+ D S GCT++EM T PW+ ++
Sbjct: 423 YWMAPEV---------VMNTNGYGL--PVDISSLGCTILEMATSKPPWSQFEGVAAIFKI 569
Query: 383 VKAKKQP--PQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML 426
+K P P++ S + I +CLQ P RPT ++L
Sbjct: 570 GNSKDMPEIPEHLS-------DDAKNFIKQCLQRDPLARPTAQSLL 686
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 65.5 bits (158), Expect = 3e-11
Identities = 57/234 (24%), Positives = 102/234 (43%), Gaps = 1/234 (0%)
Frame = +2
Query: 204 RNVCTFHGAMKVDEGLCLVMD-KCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHA 262
+NV F GA LC+V + GS+ + + G L +L+ D+++G+ LH
Sbjct: 110 KNVVQFIGACTRTPNLCIVTEFMSRGSLYDFLHKQRGVFKLPSLLKVAIDVSKGMNYLHQ 289
Query: 263 AGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSP 322
++ LK NLL+D N V+D+G+A ++ + A +
Sbjct: 290 NNIIHRDLKTGNLLMDENELVKVADFGVARVITQSGVMTAETG---------------TY 424
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
+ APE E ++D +SFG L E+ TG +P++ L+ + V
Sbjct: 425 RWMAPEVIEHKP------------YDQKADVFSFGIALWELLTGELPYSYLTPLQAAVGV 568
Query: 383 VKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQEI 436
V+ +P S+ PR L +++ C + P +RP F+ ++ I +E+
Sbjct: 569 VQKGLRP----SIPKNTHPR-LSELLQRCWKQDPIERPAFSEIIEILQNIAKEV 715
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 63.5 bits (153), Expect = 1e-10
Identities = 46/158 (29%), Positives = 72/158 (45%)
Frame = +3
Query: 237 NEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKK 296
N+G+L + +Y + V H+ GV LKP NLLLD N VSD+GL+ +
Sbjct: 702 NKGKLNEDDARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDENEDLKVSDFGLSAL--- 872
Query: 297 PSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSF 356
+ D + C +P Y APE + KK + ++D WS
Sbjct: 873 ----PEQRRDDGMLVTPCG-----TPAYVAPEVLK--KKGYD---------GSKADIWSC 992
Query: 357 GCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYAS 394
G L + +G +P+ G + IYR+ KA+ + P++ S
Sbjct: 993 GVILYALLSGYLPFQGENVMRIYRKAFKAEYEFPEWIS 1106
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 63.2 bits (152), Expect = 1e-10
Identities = 74/314 (23%), Positives = 127/314 (39%)
Frame = +2
Query: 147 RRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNV 206
R +G+G A V + G +V K + EGM +D + ++ +R N+
Sbjct: 50 RVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGM-MDQIKREISIMRLVRH--PNI 220
Query: 207 CTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVV 266
M + +M+ G + +G+L + RY + V H+ GV
Sbjct: 221 VNLKEVMATKTKIFFIMEYIRGGELFAKVA-KGKLKDDLARRYFQQLISAVDYCHSRGVS 397
Query: 267 CMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTA 326
LKP NLLLD N + VSD+GL+ + PE + + +P Y A
Sbjct: 398 HRDLKPENLLLDENENLKVSDFGLSGL----------PE--QLRQDGLLHTQCGTPAYVA 541
Query: 327 PEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAK 386
PE KK + F ++D WS G L + G +P+ + +Y +V++A+
Sbjct: 542 PEVLR--KKGYDGF---------KTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLRAE 688
Query: 387 KQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQEIPRSPPASPDN 446
Q P + S E K+I + L P++R T ++++ + P PD
Sbjct: 689 FQFPPWFS-------PESKKLISKILVADPNRRITISSIMRVSWFQKGFSASIPIPDPDE 847
Query: 447 DLVKGSVSNVTEAS 460
+++ +E S
Sbjct: 848 SNFNSDLNSSSEQS 889
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 54.3 bits (129), Expect = 6e-08
Identities = 47/164 (28%), Positives = 73/164 (43%), Gaps = 3/164 (1%)
Frame = +1
Query: 133 PVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDW---M 189
P I +G +Q L R +G G A V ++ G VAVK + ++ +D +
Sbjct: 70 PRIILGKYQ---LTRFLGRGNFAKVYQAVSLTDGTT----VAVKMIDKSKTVDASMEPRI 228
Query: 190 LGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRY 249
+ +++ +RR N+ H M + L++D G + GRL RY
Sbjct: 229 VREIDAMRRLQHH-PNILKIHEVMATRTKIYLIVDYAGGGELFSKISRRGRLPEPLARRY 405
Query: 250 GADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATI 293
+ + H GV LKP NLLLDA G+ VSD+GL+ +
Sbjct: 406 FQQLVSALCFCHRNGVAHRDLKPQNLLLDAEGNLKVSDFGLSAL 537
>TC9041 similar to UP|Q8LLW2 (Q8LLW2) Receptor-like kinase Xa21-binding
protein 3, partial (43%)
Length = 848
Score = 52.4 bits (124), Expect = 2e-07
Identities = 42/135 (31%), Positives = 63/135 (46%), Gaps = 13/135 (9%)
Frame = +1
Query: 517 LHLACRRGSAELVETILDYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNANVTSR 576
LH+A G E++ +LD VDVL++ PL+ A+ G CV LI AN+
Sbjct: 283 LHVAAVNGRIEVLSMLLDR-NVKVDVLNRHKQTPLMLAVIHGRTGCVEKLIDAGANIL-- 453
Query: 577 LRDGL-GPSVAHVCAYHGQPDCMRELLLAG-----ADP-------NAVDDEGESVLHRAI 623
+ D L + H AY G DC++ +L A AD N D G + LH A
Sbjct: 454 MFDSLRRRTCLHYAAYFGHLDCLKAILSAAHSTPVADSWGFARFVNIRDGNGATPLHLAA 633
Query: 624 AKKFTDCALVIVENG 638
++ +C +++NG
Sbjct: 634 RQRRHECLHSLLDNG 678
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 51.6 bits (122), Expect = 4e-07
Identities = 59/240 (24%), Positives = 99/240 (40%), Gaps = 10/240 (4%)
Frame = +2
Query: 142 DLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSM 201
D L+ +G G A V + AV +VAVK V DLD LED+RR +
Sbjct: 86 DYNLLEEVGYGASATV--YRAVFLPR--DEEVAVKCV------DLDRCNANLEDIRREAQ 235
Query: 202 WC-----RNVCTFHGAMKVDEGLCLVMD-----KCFGSVQSEMLRNEGRLTLEQVLRYGA 251
RNV H + VD L +VM C ++ + +L+
Sbjct: 236 TMSLIDHRNVVRAHWSFVVDRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSILK--- 406
Query: 252 DIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKI 311
+ + + LH G + +K N+LL ++G ++D+G++ + D+ +
Sbjct: 407 ETLKALEYLHRHGHIHRDVKAGNILLGSDGQVKLADFGVSASM-----------FDAGER 553
Query: 312 HSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWA 371
+ +P + APE +P G G + ++D WSFG T +E+ G P++
Sbjct: 554 QRNRNTFVGTPCWMAPEVLQP-----------GTGYNFKADVWSFGITALELAHGHAPFS 700
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 48.5 bits (114), Expect = 3e-06
Identities = 52/168 (30%), Positives = 75/168 (43%), Gaps = 6/168 (3%)
Frame = +3
Query: 216 DEGLCLVMDKCFGSVQSEMLRNEG--RLTLEQVLRYGADI--ARGVVELHAAG--VVCMS 269
DE L + GS+ + + N+G R L +R G + ARG+ LH+ G V +
Sbjct: 474 DEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQGPNVSHGN 653
Query: 270 LKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEA 329
+K SN+LL + A VSD+GLA ++ PS R Y APE
Sbjct: 654 IKASNILLTKSYEAKVSDFGLAHLV-GPSSTPNR-----------------VAGYRAPEV 779
Query: 330 WEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEE 377
+P K +S ++D +SFG L+E+ TG P L EE
Sbjct: 780 TDPRK------------VSQKADVYSFGVLLLELLTGKAPTHALLNEE 887
>AV422334
Length = 493
Score = 46.6 bits (109), Expect = 1e-05
Identities = 37/131 (28%), Positives = 56/131 (42%), Gaps = 10/131 (7%)
Frame = +1
Query: 246 VLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLA-TILKKPSCWKARP 304
V G + V +H ++ LKP N+L ++ + VV DY + K P +K P
Sbjct: 7 VRELGRQLLECVAFMHDLRLIHTDLKPENILFISSEY-VVPDYKVTFRSPKDPISFKRLP 183
Query: 305 ECDSAKI---------HSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWS 355
+ + K+ H I+ + HY APE G+G S D WS
Sbjct: 184 KSSAIKVIDFGSTAYEHQGHNYIVSTRHYRAPEVIL------------GLGWSSPCDIWS 327
Query: 356 FGCTLVEMCTG 366
GC L+E+C+G
Sbjct: 328 VGCILMELCSG 360
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 45.8 bits (107), Expect = 2e-05
Identities = 38/134 (28%), Positives = 59/134 (43%), Gaps = 3/134 (2%)
Frame = +2
Query: 239 GRLTLEQVLRYGADIARGVVELH---AAGVVCMSLKPSNLLLDANGHAVVSDYGLATILK 295
G L E + + ARG+ +H + ++ +K +N+LLDA+ A V+D+GLA L
Sbjct: 2468 GHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLY 2647
Query: 296 KPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWS 355
P ++ M I S Y APE +K + +SD +S
Sbjct: 2648 DPGASQS------------MSSIAGSYGYIAPEYAYTLK------------VDEKSDVYS 2755
Query: 356 FGCTLVEMCTGAIP 369
FG L+E+ G P
Sbjct: 2756 FGVVLLELIIGRKP 2797
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 45.8 bits (107), Expect = 2e-05
Identities = 35/131 (26%), Positives = 56/131 (42%)
Frame = +1
Query: 233 EMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLAT 292
+++R+ L+ E + I RG+ +H+A V+ LKPSNLLL+AN + D+GLA
Sbjct: 103 QIIRSNQGLSEEHCQYFLYQILRGLKYIHSANVLHRDLKPSNLLLNANCDLKICDFGLAR 282
Query: 293 ILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESD 352
+ + E + + E ++ S YTA D
Sbjct: 283 VTSETD---FMTEYVVTRWYRAPELLLNSSDYTA-----------------------AID 384
Query: 353 AWSFGCTLVEM 363
WS GC +E+
Sbjct: 385 VWSVGCIFMEL 417
>BI418653
Length = 531
Score = 45.8 bits (107), Expect = 2e-05
Identities = 31/99 (31%), Positives = 50/99 (50%)
Frame = +3
Query: 473 RLHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVETI 532
RL L +EGD+ G+R+ L S N ++ DG+TALH+A +G +V +
Sbjct: 183 RLMYLANEGDLDGIREVLESGVSVN----------FRDIDGRTALHIAACQGLTHVVALL 332
Query: 533 LDYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNA 571
L+ A VD D+ G PL A+ +++ + + K A
Sbjct: 333 LE-KGAQVDTKDRWGSTPLADAIFYKNNDVIKLMEKHGA 446
>TC9537 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 480
Score = 45.4 bits (106), Expect = 3e-05
Identities = 35/123 (28%), Positives = 57/123 (45%), Gaps = 3/123 (2%)
Frame = +1
Query: 247 LRYGADIARGVVELH---AAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKAR 303
LR +A G+ +H + +V +K SN+LLD +A V+D+GLA +L K +
Sbjct: 43 LRIAIGVAHGLCYMHHDCSPPIVHRDIKTSNILLDTGFNAKVADFGLARMLMKSGQF--- 213
Query: 304 PECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEM 363
+ M ++ S Y APE + + +S + D +SFG L+E+
Sbjct: 214 ---------NTMSAVIGSFGYMAPEYVQTTR------------VSVKVDVYSFGVVLLEL 330
Query: 364 CTG 366
TG
Sbjct: 331 ATG 339
>CN825384
Length = 729
Score = 45.1 bits (105), Expect = 4e-05
Identities = 45/167 (26%), Positives = 71/167 (41%), Gaps = 17/167 (10%)
Frame = +3
Query: 274 NLLLDANGHAVVSDYGLATI---LKKPSC--WKARPECDSAKIHSCMECIMLSPHYTAPE 328
N+LLD N ++D+GLA +K+ S WK+ + M ++ Y APE
Sbjct: 3 NILLDKNLCPHLTDFGLAEYKNDIKRVSLENWKSSGKPTGGFHKKNMVGTLV---YMAPE 173
Query: 329 AWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQ 388
++K L+ + +SD +SFG ++ E+ TG +P+ L AE V++
Sbjct: 174 I---LRKELH---------TDKSDVYSFGVSINELLTGVVPYTDLRAEAQAHTVLEMNYT 317
Query: 389 PPQYASVVGG------------GIPRELWKMIGECLQFKPSKRPTFN 423
Q V GIP L MI +C P RP+ +
Sbjct: 318 EQQLIEAVVSDGLRPVLASKDFGIPSRLLSMIQKCWDTNPKSRPSID 458
>BI417326
Length = 478
Score = 45.1 bits (105), Expect = 4e-05
Identities = 38/134 (28%), Positives = 61/134 (45%)
Frame = +2
Query: 438 RSPPASPDNDLVKGSVSNVTEASPVPELEIPQDPNRLHRLVSEGDVTGVRDFLAKAASEN 497
R +PD++ + + + V E P RL L +EGD+ G+ + L + N
Sbjct: 80 RQSSLAPDHEDSSAVAAAAADDASVTEAVDPAV--RLMYLANEGDLEGITELLDAGSDVN 253
Query: 498 ESNFISSLLEAQNADGQTALHLACRRGSAELVETILDYPEANVDVLDKDGDPPLVFALAA 557
++ DG+TALH+A ++V+ +L A+VD D+ G PL AL
Sbjct: 254 ----------FRDIDGRTALHVAACHRRTDVVDLLLQ-RGADVDTQDRWGSTPLADALYY 400
Query: 558 GSHECVCSLIKRNA 571
+H+ V L K A
Sbjct: 401 KNHDVVKLLEKHGA 442
Score = 31.2 bits (69), Expect = 0.55
Identities = 17/49 (34%), Positives = 27/49 (54%)
Frame = +2
Query: 590 AYHGQPDCMRELLLAGADPNAVDDEGESVLHRAIAKKFTDCALVIVENG 638
A G + + ELL AG+D N D +G + LH A + TD ++++ G
Sbjct: 194 ANEGDLEGITELLDAGSDVNFRDIDGRTALHVAACHRRTDVVDLLLQRG 340
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 44.7 bits (104), Expect = 5e-05
Identities = 41/142 (28%), Positives = 67/142 (46%), Gaps = 3/142 (2%)
Frame = +3
Query: 228 GSVQSEMLRNEGRLTLEQVLRYGADIARGVVELH---AAGVVCMSLKPSNLLLDANGHAV 284
GSVQ+ ++ RL + +A+G+ +H + VV LKPSN+LLD+ +A
Sbjct: 159 GSVQNNIIDWPRRLHI------AIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAK 320
Query: 285 VSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDG 344
V+D+GLA + KP + + M + + Y APE +
Sbjct: 321 VADFGLAMMSVKP------------EELATMSAVAGTFGYIAPE------------YALT 428
Query: 345 IGISPESDAWSFGCTLVEMCTG 366
I ++ + D +SFG L+E+ TG
Sbjct: 429 IRVNEKIDVYSFGVILLELTTG 494
>TC17660 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, partial (64%)
Length = 717
Score = 44.7 bits (104), Expect = 5e-05
Identities = 36/131 (27%), Positives = 59/131 (44%)
Frame = +2
Query: 233 EMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLAT 292
+++++ L+ + + + RG+ LH+A ++ LKP NLL++AN + D+GLA
Sbjct: 200 QIIKSSQSLSNDHCQYFLFQLLRGLKYLHSANILHRDLKPGNLLINANCDLKICDFGLAR 379
Query: 293 ILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESD 352
I +C K + M +++ Y APE L D G S D
Sbjct: 380 I----NCSK----------NQFMTEYVVTRWYRAPEL---------LLCCDNYGTS--ID 484
Query: 353 AWSFGCTLVEM 363
WS GC E+
Sbjct: 485 VWSVGCIFAEL 517
>BI420143
Length = 509
Score = 44.7 bits (104), Expect = 5e-05
Identities = 26/91 (28%), Positives = 49/91 (53%), Gaps = 2/91 (2%)
Frame = +2
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFG-SVQSEMLRNEGR-LTLEQVLRYGADIARGVVELHA 262
N+ F GA + C+V + G SV+ + + + R + L+ ++ D+ARG+ +H
Sbjct: 167 NIVRFIGACRKPMVWCIVTEYAKGGSVRQFLTKRQNRSVPLKLAVKQALDVARGMAYVHG 346
Query: 263 AGVVCMSLKPSNLLLDANGHAVVSDYGLATI 293
G++ LK NLL+ + ++D+G+A I
Sbjct: 347 LGLIHRDLKSDNLLIFGDKSIKIADFGVARI 439
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 44.7 bits (104), Expect = 5e-05
Identities = 43/188 (22%), Positives = 72/188 (37%), Gaps = 2/188 (1%)
Frame = +2
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGA--DIARGVVELHA 262
N+ + D+ L +V++ C + + ++ +V G + V H
Sbjct: 248 NILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHCHR 427
Query: 263 AGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSP 322
GV +KP N+L G ++D+G A W D + M ++ +P
Sbjct: 428 LGVAHRDVKPDNVLFGGGGDLKLADFGSAE-------WFG----DGRR----MSGVVGTP 562
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
+Y APE G + D WS G L M +G P+ G SA EI+ V
Sbjct: 563 YYVAPEVLM------------GREYGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAV 706
Query: 383 VKAKKQPP 390
++ + P
Sbjct: 707 IRGNLRFP 730
>TC14527 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(42%)
Length = 1095
Score = 44.3 bits (103), Expect = 6e-05
Identities = 33/124 (26%), Positives = 54/124 (42%)
Frame = +2
Query: 6 CSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNSVQALR 65
C++C ++ + + +L QCGHGF C+ S S +CP CR V V Q
Sbjct: 509 CAICLAEFDAGDEIRVLPQCGHGFHVGCIDTWLGSHS----SCPSCRQVLAVAARCQRCG 676
Query: 66 KNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNSRGSQA 125
+ L AA++++ GGG ++ E+ G+D+ NS + +
Sbjct: 677 R--------LPAANNSS---GGGATSVAV-------NEPELKSGEDENAPA-VNSNNNAS 799
Query: 126 SSSG 129
SSG
Sbjct: 800 GSSG 811
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,655,679
Number of Sequences: 28460
Number of extensions: 199124
Number of successful extensions: 1716
Number of sequences better than 10.0: 201
Number of HSP's better than 10.0 without gapping: 1555
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1658
length of query: 652
length of database: 4,897,600
effective HSP length: 96
effective length of query: 556
effective length of database: 2,165,440
effective search space: 1203984640
effective search space used: 1203984640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC126006.5


