

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.3 + phase: 0
(692 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
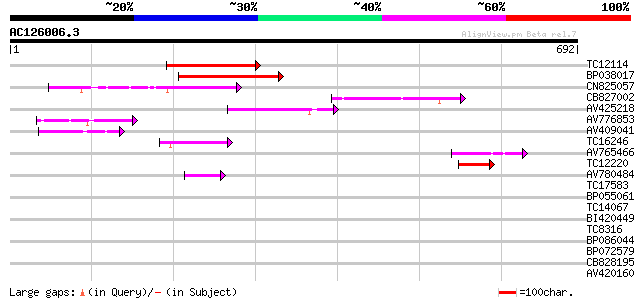
Score E
Sequences producing significant alignments: (bits) Value
TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transp... 136 1e-32
BP038017 122 1e-28
CN825057 111 3e-25
CB827002 84 6e-17
AV425218 80 1e-15
AV776853 70 1e-12
AV409041 64 6e-11
TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired re... 62 3e-10
AV765466 57 1e-08
TC12220 45 4e-05
AV780484 45 5e-05
TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired re... 40 0.001
BP055061 38 0.006
TC14067 similar to UP|O64970 (O64970) Cationic peroxidase 2 , p... 30 1.3
BI420449 30 1.3
TC8316 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Ara... 29 2.2
BP086044 29 2.9
BP072579 29 2.9
CB828195 28 3.7
AV420160 28 3.7
>TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transposase-like
{Oryza sativa (japonica cultivar-group);}, partial (8%)
Length = 488
Score = 136 bits (342), Expect = 1e-32
Identities = 62/116 (53%), Positives = 88/116 (75%), Gaps = 1/116 (0%)
Frame = +1
Query: 192 QTRRMRSLMYGEVGALLMHFKRQS-ENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSG 250
+ RR RSL++GE G +L +F+R+ EN FY+ +Q+D E++ITNVFW DA+M+ DYG G
Sbjct: 10 EKRRQRSLLHGEAGYILQYFQRKLVENSPFYHAYQLDDEDQITNVFWVDARMLIDYGYFG 189
Query: 251 DVITFDTTYMTNKDYRPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKA 306
D+++ D+TY T+ RPL VF G N+H++ V+FGA LLYDET S+ WLFE+FL+A
Sbjct: 190 DMVSLDSTYCTHSSNRPLAVFSGFNHHRKAVIFGAALLYDETTESY*WLFESFLEA 357
>BP038017
Length = 570
Score = 122 bits (307), Expect = 1e-28
Identities = 60/129 (46%), Positives = 81/129 (62%), Gaps = 1/129 (0%)
Frame = +2
Query: 207 LLMHFKR-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDY 265
LL +FK+ Q +NP FYY Q+D E ++ NVFWADA+ + Y GD + FDT Y N+
Sbjct: 179 LLNYFKKMQGKNPGFYYAIQLDDENRMINVFWADARSRSAYNYFGDAVIFDTMYRPNQYQ 358
Query: 266 RPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAK 325
P F G+N+H Q V+FG LL DE+ SF WLF T+L AM P ++ TDQD A+
Sbjct: 359 VPFAPFTGVNHHGQNVLFGCALLLDESESSFTWLFRTWLSAMNDRPPVSITTDQDRAIQA 538
Query: 326 AISVVMPQT 334
A++ V P+T
Sbjct: 539 AVAQVFPET 565
>CN825057
Length = 721
Score = 111 bits (278), Expect = 3e-25
Identities = 75/251 (29%), Positives = 129/251 (50%), Gaps = 16/251 (6%)
Frame = +1
Query: 48 AYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFK-----EGKRGVDKRDHLTKEG 102
AY FY +Y++ +GF + +SKK +F C + E G ++R + K
Sbjct: 1 AYSFYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYGVTPESDGGSNRRSSVKK-- 174
Query: 103 RVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFHTPRSHRQI--SESRACQ 160
T C A M + + GK+ + +F+ +HNH L P +H R HR + +E
Sbjct: 175 -----TDCKACMHVKR-KPDGKWIIHEFIKEHNHELLPALAYHF-RIHRNVKLAEKNNMD 333
Query: 161 VVAADESRLKRKDFQEYVFKQDGGIDDVGYL--------QTRRMRSLMYGEVGALLMHFK 212
++ A R RK + E + +Q GG ++ L + + ++ G+ +L +FK
Sbjct: 334 ILHAVSERT-RKMYVE-MSRQSGGCLNIESLVGDLNDQFKKGQYLAMDEGDAQVMLEYFK 507
Query: 213 R-QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNKDYRPLGVF 271
Q ENP+F+Y ++ E+++ N+FW DA+ INDY DV++FDT+Y+ + + P F
Sbjct: 508 HIQKENPNFFYSIDLNEEQRLRNIFWIDAKSINDYLSFNDVVSFDTSYIKSNEKLPFAPF 687
Query: 272 VGLNNHKQMVV 282
VG+N+H Q ++
Sbjct: 688 VGVNHHCQPIL 720
>CB827002
Length = 534
Score = 84.3 bits (207), Expect = 6e-17
Identities = 53/169 (31%), Positives = 84/169 (49%), Gaps = 5/169 (2%)
Frame = +1
Query: 393 EDSWLRMIFQLKEKWAWVYVRKHFTAGMRSTQLSESFNAELKNYLKSDLNLVQFFSHFGR 452
+ WL + ++ A V++R F A M TQ S+S N+ Y+ + NL QFF + +
Sbjct: 31 DHEWLSLYSSCRQG-APVHLRDTFFAEMSITQRSDSMNSYFDGYVNASTNLNQFFKLYEK 207
Query: 453 IVHGIRNNESEADYESRHKLPKLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYLGTCVKN 512
+ E ADY++ + LP L+ +PM QA +YT K F FQEE L
Sbjct: 208 ALESRNEKEVRADYDTMNTLPVLRTP-SPMEKQASELYTRKIFMRFQEELVGTLTFMASK 384
Query: 513 LKEGLYVVTNY-----DNNKERMVIGNLMDQKVACDCRKFETHGILCSH 556
+ V+T + +++K V N+++ K +C C+ FE G+LC H
Sbjct: 385 ADDDGEVITYHVAKFGEDHKAYYVKFNVLEMKASCSCQMFEFSGLLCRH 531
>AV425218
Length = 419
Score = 80.1 bits (196), Expect = 1e-15
Identities = 45/140 (32%), Positives = 74/140 (52%), Gaps = 4/140 (2%)
Frame = +2
Query: 266 RPLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAK 325
RPL VG+N+H Q V+FG LL E SF WLF++ L M G P+ ++TD EAM K
Sbjct: 2 RPLATLVGVNHHGQSVLFGCALLSSEDSESFVWLFQSLLHCMSGVPPQGIITDHSEAMKK 181
Query: 326 AISVVMPQTFHGLC-TWRIRENAQTHVNHLYQKSSKFCSD---FEACIDLHEEEGEFLNS 381
AI V+P T H C ++ +++ Q + + +S + ++A + EF +
Sbjct: 182 AIETVLPSTRHRWCLSYIMKKLPQKLLGYAQYESIRHHLQNVVYDAVV-----IDEFERN 346
Query: 382 WNVLLVEHNVSEDSWLRMIF 401
W ++ + + ++ WL +F
Sbjct: 347 WKKIVEDFGLEDNEWLNELF 406
>AV776853
Length = 625
Score = 69.7 bits (169), Expect = 1e-12
Identities = 46/131 (35%), Positives = 69/131 (52%), Gaps = 7/131 (5%)
Frame = +1
Query: 33 DYIPHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGV 92
D I HL+ F SE AY+FY Y++ GF +R++ + G + R+F C ++G R
Sbjct: 223 DDIRHLD--FGSEEEAYQFYQAYAKYQGFIVRKDDIGRDYH-GNVNMRQFVCNRQGLRSK 393
Query: 93 D-------KRDHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEPPGYFH 145
KRDH + T T C A++ + LD KIGK+KVV F HNH L P + H
Sbjct: 394 KHYNRTDRKRDH-----KPVTHTNCLAKLRVHLDYKIGKWKVVSFEECHNHELTPARFVH 558
Query: 146 TPRSHRQISES 156
+R ++++
Sbjct: 559 FIPPYRVMNDA 591
>AV409041
Length = 349
Score = 64.3 bits (155), Expect = 6e-11
Identities = 35/105 (33%), Positives = 54/105 (51%)
Frame = +3
Query: 36 PHLEMEFESEAAAYEFYNKYSRRIGFGIRREYGNKSKKDGILTSRRFTCFKEGKRGVDKR 95
PH ++EFES AAY FY +Y++ GFG + +S+ +F+C + G + ++
Sbjct: 33 PHYDIEFESHEAAYAFYKEYAKSAGFGTAKLSSRRSRASKEFIDAKFSCIRYGNK---QQ 203
Query: 96 DHLTKEGRVETRTGCDARMVISLDRKIGKYKVVDFVAQHNHLLEP 140
R + GC A M + R+ GK+ V FV +HNH L P
Sbjct: 204 SDDAINPRPSPKIGCKASMHVK-RRQDGKWYVYSFVKEHNHELLP 335
>TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired response
protein-like, partial (4%)
Length = 660
Score = 62.0 bits (149), Expect = 3e-10
Identities = 33/99 (33%), Positives = 52/99 (52%), Gaps = 9/99 (9%)
Frame = +2
Query: 183 GGIDDVGYLQTR--------RMRSLMYGEVGALLMHFKRQSEN-PSFYYDFQMDVEEKIT 233
GG + +G+ +T + + G+ A L + + +++N P F+Y F +E +
Sbjct: 65 GGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKADNDPMFFYKFTKTGDESLE 244
Query: 234 NVFWADAQMINDYGCSGDVITFDTTYMTNKDYRPLGVFV 272
N+FW D DY GDVI FD+TY NK +PL VF+
Sbjct: 245 NLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFL 361
>AV765466
Length = 599
Score = 56.6 bits (135), Expect = 1e-08
Identities = 32/93 (34%), Positives = 48/93 (51%)
Frame = -2
Query: 540 VACDCRKFETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLKGQHIELDT 599
V CDCR FE GILC H+L +L +K +P Y+L RW ++ R + +K +
Sbjct: 598 VKCDCRLFEFRGILCRHSLAILSQERVKEVPDRYVLDRWRKNMRRKYVY-VKTSYCVQHL 422
Query: 600 KAHFMRRYNDLCPQMIKLINKASKSHETYTFLS 632
K M R LC Q + A++ ET +F++
Sbjct: 421 KPQ-MERLELLCNQFKSIAVSAAEFEETSSFVN 326
>TC12220
Length = 473
Score = 45.1 bits (105), Expect = 4e-05
Identities = 18/44 (40%), Positives = 28/44 (62%)
Frame = +1
Query: 548 ETHGILCSHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHDLK 591
E G+LC H L+V ++ ++ +P YIL RWTR+A G D++
Sbjct: 1 EYEGVLCRHPLRVFXILELREVPSRYILHRWTRNAEDGVFPDIE 132
>AV780484
Length = 529
Score = 44.7 bits (104), Expect = 5e-05
Identities = 19/50 (38%), Positives = 29/50 (58%)
Frame = -1
Query: 214 QSENPSFYYDFQMDVEEKITNVFWADAQMINDYGCSGDVITFDTTYMTNK 263
Q ENP+F+Y +D+ +T+VFW D + DY S ++ T Y+ NK
Sbjct: 175 QGENPNFFYAIDLDLNRHLTSVFWVDIKGRLDYETSMMLVLIHTHYLKNK 26
>TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein, mutator-like transposase-like protein,
phytochrome A signaling protein-like, partial (3%)
Length = 666
Score = 40.4 bits (93), Expect = 0.001
Identities = 18/30 (60%), Positives = 21/30 (70%)
Frame = +3
Query: 241 QMINDYGCSGDVITFDTTYMTNKDYRPLGV 270
+MI DY GDV++ DTTY TN YRPL V
Sbjct: 3 RMIMDYEYFGDVVSLDTTYSTNNAYRPLAV 92
>BP055061
Length = 544
Score = 37.7 bits (86), Expect = 0.006
Identities = 17/55 (30%), Positives = 32/55 (57%)
Frame = -2
Query: 607 YNDLCPQMIKLINKASKSHETYTFLSKVFEESDKIIDDMLAKKFVGVESLEMSHV 661
Y DLCP+++KL +AS+S E Y F +K ++ + ++ +L K + + S +
Sbjct: 486 YKDLCPRLVKLSARASESVEAYQFTAKQLDKVMEGVEKILTSKVEEAQVITSSSI 322
>TC14067 similar to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(18%)
Length = 476
Score = 30.0 bits (66), Expect = 1.3
Identities = 15/56 (26%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Frame = +2
Query: 555 SHALKVLDVMNIKLIPQHYILKRWTRDARLGSNHD-LKGQHIELDTKAHFMRRYND 609
+HA + + PQ Y+ K W+R + L S +D L G E+ + H ++++
Sbjct: 17 AHAQRTKPYLKKMARPQDYVFKEWSRASTLLSEYDPLTGPKAEIRNQCHVAYQHHE 184
>BI420449
Length = 580
Score = 30.0 bits (66), Expect = 1.3
Identities = 17/67 (25%), Positives = 31/67 (45%)
Frame = +3
Query: 624 SHETYTFLSKVFEESDKIIDDMLAKKFVGVESLEMSHVSIPVAMEKIDNNVHTLDVGGEQ 683
S E + L++ EESDK+ D +A VE++ S +E + + ++
Sbjct: 231 SKEEFEALNRKVEESDKLADMKVAAAKAQVEAVRASENESIKRLETTQKEIDYMKTATQE 410
Query: 684 GIKKARL 690
+KKA +
Sbjct: 411 ALKKAEM 431
>TC8316 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Arabidopsis
thaliana;}, partial (63%)
Length = 1796
Score = 29.3 bits (64), Expect = 2.2
Identities = 17/64 (26%), Positives = 29/64 (44%)
Frame = +2
Query: 267 PLGVFVGLNNHKQMVVFGATLLYDETIPSFQWLFETFLKAMGGEKPKTLLTDQDEAMAKA 326
P G + L+NH +M VF L I S+ W+ + + E P L + + + K
Sbjct: 596 PPGGVIPLHNHPEMTVFSKLLFGTMHIKSYDWVVD-----LPSEMPTILKSSEIQTQEKR 760
Query: 327 ISVV 330
++ V
Sbjct: 761 LAKV 772
>BP086044
Length = 322
Score = 28.9 bits (63), Expect = 2.9
Identities = 11/33 (33%), Positives = 20/33 (60%)
Frame = +1
Query: 474 KLKMKRAPMLVQAGNIYTPKTFEEFQEEYEEYL 506
K K K+ L+ G+ Y PK+ + FQ+ +++L
Sbjct: 61 KKKKKKTKYLLCTGHFYVPKSLQNFQDTAKQFL 159
>BP072579
Length = 430
Score = 28.9 bits (63), Expect = 2.9
Identities = 20/50 (40%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Frame = +3
Query: 4 ATIPIVEGVEKL---GNNIDVNIVQNGEESVVDYIPHLEMEFESEAAAYE 50
A PIVEGVEK+ G + ++ NG+E H E E ESE + E
Sbjct: 297 AATPIVEGVEKISLTGTPLSKSVEGNGDE------VHSESESESENSESE 428
>CB828195
Length = 551
Score = 28.5 bits (62), Expect = 3.7
Identities = 29/127 (22%), Positives = 49/127 (37%), Gaps = 10/127 (7%)
Frame = +3
Query: 443 LVQFFSHFGRIVHGIRNNESEADYESRH-----KLPKLKMKRAPMLVQAGNIYTPKTFEE 497
+++FFS+F +R E +DYE H K ++ ++ + K EE
Sbjct: 159 ILKFFSNFWCSNSEVRGEEKVSDYEFNHQENESKSEGIQRDEVKKVMAELGFFCSKESEE 338
Query: 498 FQEEY-----EEYLGTCVKNLKEGLYVVTNYDNNKERMVIGNLMDQKVACDCRKFETHGI 552
+E+Y E +L+E +D N++ + D R E H +
Sbjct: 339 LEEKYGSKELSELFEDQEPSLEEVKQAFDVFDENRDGFI-----------DAR--ELHRV 479
Query: 553 LCSHALK 559
LC LK
Sbjct: 480 LCVLGLK 500
>AV420160
Length = 417
Score = 28.5 bits (62), Expect = 3.7
Identities = 22/91 (24%), Positives = 37/91 (40%)
Frame = +1
Query: 68 GNKSKKDGILTSRRFTCFKEGKRGVDKRDHLTKEGRVETRTGCDARMVISLDRKIGKYKV 127
G+KSK + S + K G++ + + R + GC AR + R +
Sbjct: 121 GSKSKGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTKLM 300
Query: 128 VDFVAQHNHLLEPPGYFHTPRSHRQISESRA 158
V + +HNH L P+SH + + A
Sbjct: 301 VTYSYEHNHSLP------VPKSHSSAASATA 375
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,534,246
Number of Sequences: 28460
Number of extensions: 158292
Number of successful extensions: 675
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 664
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 668
length of query: 692
length of database: 4,897,600
effective HSP length: 97
effective length of query: 595
effective length of database: 2,136,980
effective search space: 1271503100
effective search space used: 1271503100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC126006.3


