

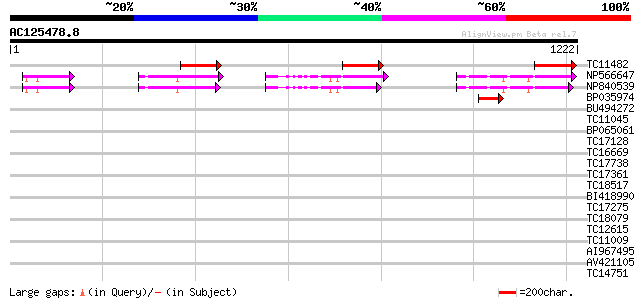
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.8 + phase: 0
(1222 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11482 similar to UP|Q8H6W8 (Q8H6W8) Pathogen-induced calmoduli... 161 5e-40
NP566647 unnamed protein product [Lotus japonicus] 108 5e-24
NP840539 hypothetical protein [Lotus corniculatus var. japonicus] 106 3e-23
BP035974 59 4e-09
BU494272 37 0.025
TC11045 35 0.071
BP065061 33 0.35
TC17128 homologue to PIR|T06431|T06431 ribosomal protein L27-5 -... 33 0.35
TC16669 similar to UP|Q9SLT9 (Q9SLT9) ZCF37 protein (T30E16.15),... 32 0.79
TC17738 similar to UP|O48679 (O48679) F3I6.5 protein, partial (8%) 31 1.0
TC17361 31 1.3
TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial ... 30 1.8
BI418990 30 2.3
TC17275 30 2.3
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 29 3.9
TC12615 homologue to AAR23711 (AAR23711) At1g55090, partial (7%) 29 3.9
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 29 5.1
AI967495 29 5.1
AV421105 28 6.7
TC14751 similar to UP|Q948T9 (Q948T9) Respiratory burst oxidase ... 28 8.7
>TC11482 similar to UP|Q8H6W8 (Q8H6W8) Pathogen-induced calmodulin-binding
protein (Fragment), partial (51%)
Length = 604
Score = 161 bits (408), Expect = 5e-40
Identities = 80/91 (87%), Positives = 84/91 (91%)
Frame = +2
Query: 1131 LKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYA 1190
LKKV+LLRRFIKALEKVRKFNPR PRYLP+EPDSE EKV LRHQDM RKG EEWMLDYA
Sbjct: 2 LKKVILLRRFIKALEKVRKFNPRGPRYLPIEPDSEAEKVLLRHQDMEARKGQEEWMLDYA 181
Query: 1191 LRQVVSKLTPARKRKVELLVEAFETVVPTVK 1221
LRQVVSKLTPAR+RKVELLVEAFETV PT+K
Sbjct: 182 LRQVVSKLTPARRRKVELLVEAFETVTPTIK 274
Score = 117 bits (294), Expect = 8e-27
Identities = 59/88 (67%), Positives = 70/88 (79%)
Frame = +2
Query: 369 LKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYA 428
LKKVILL+RF+KALEKVR N R PR LP + + EAEKVLL Q E RK EEWMLDYA
Sbjct: 2 LKKVILLRRFIKALEKVRKFNPRGPRYLPIEPDSEAEKVLLRHQDMEARKGQEEWMLDYA 181
Query: 429 LQKVISKLAPAQRQRVTLLVEAFETIRP 456
L++V+SKL PA+R++V LLVEAFET+ P
Sbjct: 182 LRQVVSKLTPARRRKVELLVEAFETVTP 265
Score = 113 bits (282), Expect = 2e-25
Identities = 54/88 (61%), Positives = 69/88 (78%)
Frame = +2
Query: 717 LKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYA 776
LKK+I+L+RF+KAL+KVR NPR PR LP + + E EKV L Q E RK EEWMLDYA
Sbjct: 2 LKKVILLRRFIKALEKVRKFNPRGPRYLPIEPDSEAEKVLLRHQDMEARKGQEEWMLDYA 181
Query: 777 LQKVISKLAPAQRQRVTLLIEAFETLRP 804
L++V+SKL PA+R++V LL+EAFET+ P
Sbjct: 182 LRQVVSKLTPARRRKVELLVEAFETVTP 265
>NP566647 unnamed protein product [Lotus japonicus]
Length = 2205
Score = 108 bits (270), Expect = 5e-24
Identities = 90/279 (32%), Positives = 139/279 (49%), Gaps = 20/279 (7%)
Frame = +1
Query: 963 EAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTRE 1022
E AV E KN + + G K ++ D+ E N++ + +++ +++ +T E
Sbjct: 1354 EVALQAVKEEKNTHFEAQTHGT-----KSVLEDI-EFNTQETDHLSNAASHEHDQSSTEE 1515
Query: 1023 ISVSYENTPVVIQDMPFKDRAVVDAEV----ELRQIEAIKMVED-------AIDSILPDT 1071
+ + NT +D + +D EV ++ +AI+ E AID D+
Sbjct: 1516 VFEHFTNT----RDNNRESEKHMDNEVSCASKVLDEDAIENCEGHTNSETFAIDESCEDS 1683
Query: 1072 QPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVEERK---EESVSKE------VNKPNQ 1122
P S G+ E L + + + I+++++ E+ V V+ Q
Sbjct: 1684 NP----SLEINDEGLSQENLINLSAEPKESSIIIQDQELLEEDQVRVSRFHTSCVDSEQQ 1851
Query: 1123 KLSRNWSNLKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGT 1182
+NW K V +R + E+VR+ NPR+P +LPL PD E EKV L+HQ + ERK
Sbjct: 1852 NTGKNW---KWAVRHKRPDQDNEEVRRINPRKPNFLPLNPDPEPEKVDLKHQMIDERKHA 2022
Query: 1183 EEWMLDYALRQVVSKLTPARKRKVELLVEAFETVVPTVK 1221
+EWMLD+ALRQ V+KL PA K KV LLVEAFETV+ K
Sbjct: 2023 DEWMLDFALRQAVTKLVPAGKMKVALLVEAFETVMSIPK 2139
Score = 97.1 bits (240), Expect = 2e-20
Identities = 54/126 (42%), Positives = 75/126 (58%), Gaps = 15/126 (11%)
Frame = +1
Query: 28 GIDGNNR-----NSTSDAGNKSQRVMTRRLSLKPVR-------ISAKKPSLH---KATCS 72
G DG N +++S + K + +TR +LKP I+ K+ ++ KATCS
Sbjct: 244 GSDGKNLPQKCLSNSSVSSKKPSKTLTRSSTLKPCSGYPIKSTIAVKQEDVNPQEKATCS 423
Query: 73 STIKDSHFPNHIDLPQEGSSSQGVSAVKVCTYAYCSLHGHHHGDLPPLKRFVSMRRRQLK 132
ST+KDS FP ++ L G+ S+G S +KVC Y YCSL+ HHH LP L F+S RRR L+
Sbjct: 424 STLKDSKFPTYLMLNPGGTESEGTSVMKVCRYTYCSLNSHHHARLPQLNSFMSARRRLLE 603
Query: 133 SQKSTK 138
+QKS K
Sbjct: 604 TQKSVK 621
Score = 94.0 bits (232), Expect = 1e-19
Identities = 83/300 (27%), Positives = 132/300 (43%), Gaps = 35/300 (11%)
Frame = +1
Query: 552 QEKSIVKEDLKHGTSTT--DVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGK 609
+EK+ E HGT + D+ + QE D LSN +++
Sbjct: 1378 EEKNTHFEAQTHGTKSVLEDIEFNTQETDH--------------LSNAASHEH------- 1494
Query: 610 DKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDD 669
D+ E+ VF N+ +N E++ MD E + K DE + E
Sbjct: 1495 DQSSTEE---VFEHFTNTRDNN-RESEKHMDNE----VSCASKVLDEDAIENCEG----- 1635
Query: 670 RSKSRSYGSDELLEKSEGERE-------EMNATSFTETPKEAK----------------- 705
+ S ++ DE E S E + N + + PKE+
Sbjct: 1636 HTNSETFAIDESCEDSNPSLEINDEGLSQENLINLSAEPKESSIIIQDQELLEEDQVRVS 1815
Query: 706 ---------KTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVF 756
+ +N K+W K + KR + ++VR INPR+P LP + + E EKV
Sbjct: 1816 RFHTSCVDSEQQNTGKNW---KWAVRHKRPDQDNEEVRRINPRKPNFLPLNPDPEPEKVD 1986
Query: 757 LNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQDAENGLRSSA 816
L Q +ERK ++EWMLD+AL++ ++KL PA + +V LL+EAFET+ I E +R+++
Sbjct: 1987 LKHQMIDERKHADEWMLDFALRQAVTKLVPAGKMKVALLVEAFETVMSIPKCEAHIRNNS 2166
Score = 85.9 bits (211), Expect = 4e-17
Identities = 62/208 (29%), Positives = 96/208 (45%), Gaps = 24/208 (11%)
Frame = +1
Query: 278 NSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVED-LSSEGHSKSRGNETDEVLLEK 336
N+ N E++ MD+E + K DE + E +SE + E LE
Sbjct: 1534 NTRDNNRESEKHMDNE----VSCASKVLDEDAIENCEGHTNSETFAIDESCEDSNPSLEI 1701
Query: 337 SGGKIEERNTTTFTESPKEVPKM-----------------------ESKQKSWSHLKKVI 373
+ + + N + PKE + +Q + + K +
Sbjct: 1702 NDEGLSQENLINLSAEPKESSIIIQDQELLEEDQVRVSRFHTSCVDSEQQNTGKNWKWAV 1881
Query: 374 LLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVI 433
KR + E+VR IN R+P LP + + E EKV L Q +ERK ++EWMLD+AL++ +
Sbjct: 1882 RHKRPDQDNEEVRRINPRKPNFLPLNPDPEPEKVDLKHQMIDERKHADEWMLDFALRQAV 2061
Query: 434 SKLAPAQRQRVTLLVEAFETIRPVQDAE 461
+KL PA + +V LLVEAFET+ + E
Sbjct: 2062 TKLVPAGKMKVALLVEAFETVMSIPKCE 2145
>NP840539 hypothetical protein [Lotus corniculatus var. japonicus]
Length = 3654
Score = 106 bits (264), Expect = 3e-23
Identities = 88/273 (32%), Positives = 136/273 (49%), Gaps = 20/273 (7%)
Frame = +1
Query: 963 EAPTSAVVESKNQLEKQGSTGLWFTVFKHMVSDMTENNSKTSTDVADEKDSKYEDITTRE 1022
E AV E KN + + G K ++ D+ E N++ + +++ +++ +T E
Sbjct: 2806 EVALQAVKEEKNTHFEAQTHGT-----KSVLEDI-EFNTQETDHLSNAASHEHDQSSTEE 2967
Query: 1023 ISVSYENTPVVIQDMPFKDRAVVDAEV----ELRQIEAIKMVED-------AIDSILPDT 1071
+ + NT +D + +D EV ++ +AI+ E AID D+
Sbjct: 2968 VFEHFTNT----RDNNRESEKHMDNEVSCASKVLDEDAIENCEGHTNSETFAIDESCEDS 3135
Query: 1072 QPLPDNSTIDRTGGIYSEGLNQKEQKMESGNGIVEERK---EESVSKE------VNKPNQ 1122
P S G+ E L + + + I+++++ E+ V V+ Q
Sbjct: 3136 NP----SLEINDEGLSQENLINLSAEPKESSIIIQDQELLEEDQVRVSRFHTSCVDSEQQ 3303
Query: 1123 KLSRNWSNLKKVVLLRRFIKALEKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGT 1182
+NW K V +R + E+VR+ NPR+P +LPL PD E EKV L+HQ + ERK
Sbjct: 3304 NTGKNW---KWAVRHKRPDQDNEEVRRINPRKPNFLPLNPDPEPEKVDLKHQMIDERKHA 3474
Query: 1183 EEWMLDYALRQVVSKLTPARKRKVELLVEAFET 1215
+EWMLD+ALRQ V+KL PA K KV LLVEAFET
Sbjct: 3475 DEWMLDFALRQAVTKLVPAGKMKVALLVEAFET 3573
Score = 97.1 bits (240), Expect = 2e-20
Identities = 54/126 (42%), Positives = 75/126 (58%), Gaps = 15/126 (11%)
Frame = +1
Query: 28 GIDGNNR-----NSTSDAGNKSQRVMTRRLSLKPVR-------ISAKKPSLH---KATCS 72
G DG N +++S + K + +TR +LKP I+ K+ ++ KATCS
Sbjct: 1696 GSDGKNLPQKCLSNSSVSSKKPSKTLTRSSTLKPCSGYPIKSTIAVKQEDVNPQEKATCS 1875
Query: 73 STIKDSHFPNHIDLPQEGSSSQGVSAVKVCTYAYCSLHGHHHGDLPPLKRFVSMRRRQLK 132
ST+KDS FP ++ L G+ S+G S +KVC Y YCSL+ HHH LP L F+S RRR L+
Sbjct: 1876 STLKDSKFPTYLMLNPGGTESEGTSVMKVCRYTYCSLNSHHHARLPQLNSFMSARRRLLE 2055
Query: 133 SQKSTK 138
+QKS K
Sbjct: 2056 TQKSVK 2073
Score = 89.4 bits (220), Expect = 3e-18
Identities = 80/285 (28%), Positives = 124/285 (43%), Gaps = 35/285 (12%)
Frame = +1
Query: 552 QEKSIVKEDLKHGTSTT--DVPYGVQERDQKYIKKWHLMYKQAVLSNTGKYDNKLPVVGK 609
+EK+ E HGT + D+ + QE D LSN +++
Sbjct: 2830 EEKNTHFEAQTHGTKSVLEDIEFNTQETDH--------------LSNAASHEH------- 2946
Query: 610 DKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDD 669
D+ E+ VF N+ +N E++ MD E + K DE + E
Sbjct: 2947 DQSSTEE---VFEHFTNTRDNN-RESEKHMDNE----VSCASKVLDEDAIENCEG----- 3087
Query: 670 RSKSRSYGSDELLEKSEGERE-------EMNATSFTETPKEAK----------------- 705
+ S ++ DE E S E + N + + PKE+
Sbjct: 3088 HTNSETFAIDESCEDSNPSLEINDEGLSQENLINLSAEPKESSIIIQDQELLEEDQVRVS 3267
Query: 706 ---------KTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPRELPSDANFEGEKVF 756
+ +N K+W K + KR + ++VR INPR+P LP + + E EKV
Sbjct: 3268 RFHTSCVDSEQQNTGKNW---KWAVRHKRPDQDNEEVRRINPRKPNFLPLNPDPEPEKVD 3438
Query: 757 LNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFET 801
L Q +ERK ++EWMLD+AL++ ++KL PA + +V LL+EAFET
Sbjct: 3439 LKHQMIDERKHADEWMLDFALRQAVTKLVPAGKMKVALLVEAFET 3573
Score = 83.2 bits (204), Expect = 2e-16
Identities = 61/200 (30%), Positives = 93/200 (46%), Gaps = 24/200 (12%)
Frame = +1
Query: 278 NSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVED-LSSEGHSKSRGNETDEVLLEK 336
N+ N E++ MD+E + K DE + E +SE + E LE
Sbjct: 2986 NTRDNNRESEKHMDNE----VSCASKVLDEDAIENCEGHTNSETFAIDESCEDSNPSLEI 3153
Query: 337 SGGKIEERNTTTFTESPKEVPKM-----------------------ESKQKSWSHLKKVI 373
+ + + N + PKE + +Q + + K +
Sbjct: 3154 NDEGLSQENLINLSAEPKESSIIIQDQELLEEDQVRVSRFHTSCVDSEQQNTGKNWKWAV 3333
Query: 374 LLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVI 433
KR + E+VR IN R+P LP + + E EKV L Q +ERK ++EWMLD+AL++ +
Sbjct: 3334 RHKRPDQDNEEVRRINPRKPNFLPLNPDPEPEKVDLKHQMIDERKHADEWMLDFALRQAV 3513
Query: 434 SKLAPAQRQRVTLLVEAFET 453
+KL PA + +V LLVEAFET
Sbjct: 3514 TKLVPAGKMKVALLVEAFET 3573
>BP035974
Length = 162
Score = 59.3 bits (142), Expect = 4e-09
Identities = 33/54 (61%), Positives = 40/54 (73%)
Frame = +3
Query: 1011 KDSKYEDITTREISVSYENTPVVIQDMPFKDRAVVDAEVELRQIEAIKMVEDAI 1064
K+S E T+ SVSYEN PV QDM FK++ V D ++ELRQIEAIKMVE+AI
Sbjct: 3 KESVDEGSRTQGTSVSYENKPVTNQDMHFKEQ-VADRDIELRQIEAIKMVEEAI 161
>BU494272
Length = 469
Score = 36.6 bits (83), Expect = 0.025
Identities = 17/25 (68%), Positives = 20/25 (80%)
Frame = +3
Query: 1197 KLTPARKRKVELLVEAFETVVPTVK 1221
+L PARK+KV LLVEAFE V+P K
Sbjct: 12 ELAPARKKKVALLVEAFEAVMPNPK 86
Score = 34.7 bits (78), Expect = 0.093
Identities = 15/34 (44%), Positives = 25/34 (73%)
Frame = +3
Query: 783 KLAPAQRQRVTLLIEAFETLRPIQDAENGLRSSA 816
+LAPA++++V LL+EAFE + P + LR+S+
Sbjct: 12 ELAPARKKKVALLVEAFEAVMPNPKCDTRLRNSS 113
Score = 32.0 bits (71), Expect = 0.60
Identities = 13/22 (59%), Positives = 19/22 (86%)
Frame = +3
Query: 435 KLAPAQRQRVTLLVEAFETIRP 456
+LAPA++++V LLVEAFE + P
Sbjct: 12 ELAPARKKKVALLVEAFEAVMP 77
>TC11045
Length = 507
Score = 35.0 bits (79), Expect = 0.071
Identities = 16/25 (64%), Positives = 21/25 (84%)
Frame = -3
Query: 908 GGHGEQFSVTKSLILNGIVRSLRSN 932
G +GE S++KS ILNGIV+SLR+N
Sbjct: 298 GDNGEPLSMSKSSILNGIVKSLRNN 224
>BP065061
Length = 302
Score = 32.7 bits (73), Expect = 0.35
Identities = 19/68 (27%), Positives = 34/68 (49%), Gaps = 2/68 (2%)
Frame = +2
Query: 112 HHHGD--LPPLKRFVSMRRRQLKSQKSTKKDGRSKQVGNARKGTQKTKTVHSEDGNSQQN 169
+ HG+ LP ++ Q+++Q T + G+ K ++ KTK +HS NSQQ
Sbjct: 14 NEHGEQPLPSEQK*TGTAPLQIRTQTETXQKGKRKTKEPXKETATKTKRLHSRS*NSQQK 193
Query: 170 VKNVSMES 177
S+++
Sbjct: 194 RTQSSLQT 217
>TC17128 homologue to PIR|T06431|T06431 ribosomal protein L27-5 - garden pea
{Pisum sativum;}, complete
Length = 576
Score = 32.7 bits (73), Expect = 0.35
Identities = 16/40 (40%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Frame = +2
Query: 949 DIKDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTG--LWF 986
D+KDVV D L+ + +A+ E+K +LE++ TG WF
Sbjct: 293 DLKDVVVPDSLQSKDKKVAALKETKKRLEERFKTGKNRWF 412
>TC16669 similar to UP|Q9SLT9 (Q9SLT9) ZCF37 protein (T30E16.15), partial
(13%)
Length = 667
Score = 31.6 bits (70), Expect = 0.79
Identities = 29/115 (25%), Positives = 52/115 (45%), Gaps = 11/115 (9%)
Frame = +3
Query: 261 PLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNV--------IELVQKAFDEI--LL 310
P+ K KE +++ + +SE D+D +++ V I V+ A+ +
Sbjct: 258 PVKSKRKENRDKNPFSSRGLDKFSELLEDLDQKRQKVYSRMNPHDISFVRFAYTSSNDFV 437
Query: 311 PEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEERNTTTFTESPKEV-PKMESKQK 364
P V + + H K +GNE +V S + E+ E K+ PK+ES++K
Sbjct: 438 PIVVKVKNRDHKK-QGNEEFKVRHLTSFSESMEKTAAAAVEERKQQHPKLESEKK 599
>TC17738 similar to UP|O48679 (O48679) F3I6.5 protein, partial (8%)
Length = 599
Score = 31.2 bits (69), Expect = 1.0
Identities = 27/96 (28%), Positives = 39/96 (40%), Gaps = 20/96 (20%)
Frame = -1
Query: 125 SMRRRQLKSQKSTKKDGRSKQVGNARKGTQKTKTVH-------------SEDGNSQQNVK 171
S +RRQ S S + R+ V R+G + +KTVH + G +Q+
Sbjct: 302 SKQRRQGNSDHSPQPPPRTSPVSGDRRGQRGSKTVHRNPASKP*PSDQRTHSGENQRR*S 123
Query: 172 N-VSMESSPFKPHD------APPSTVNECDTSTKDK 200
N VS P PH PPS + + T+ K
Sbjct: 122 NTVSPSRGPSGPHSPPCGSHCPPSPTPKSNDRTRRK 15
>TC17361
Length = 398
Score = 30.8 bits (68), Expect = 1.3
Identities = 14/23 (60%), Positives = 17/23 (73%)
Frame = -3
Query: 908 GGHGEQFSVTKSLILNGIVRSLR 930
G GE S++KS ILNG VRSL+
Sbjct: 228 GDSGEPLSMSKSFILNGFVRSLK 160
>TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (4%)
Length = 493
Score = 30.4 bits (67), Expect = 1.8
Identities = 23/93 (24%), Positives = 45/93 (47%)
Frame = -2
Query: 249 VLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEI 308
V+ G+ +++ E+E+E GEED E ++ + + D+E+ + + V + E+
Sbjct: 420 VIDGAGEDNDRAVAEEEEEEPGEEDGEEDDDGDGVPDEGDEEDEEEDHGV--VAEEVAEV 247
Query: 309 LLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKI 341
+L E +EG G E +E + G+I
Sbjct: 246 VL-HAEGGFAEGVRAGEGVEGEEFFPGAARGEI 151
>BI418990
Length = 454
Score = 30.0 bits (66), Expect = 2.3
Identities = 17/60 (28%), Positives = 29/60 (48%)
Frame = +3
Query: 277 NNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETDEVLLEK 336
NNS RN S+ +S D + + L Q A+D +L +++ S H+ + L+ K
Sbjct: 27 NNSLRNSSQFNSACDSAFTHCLSLTQHAYDGVLPYQLKTASDHIHTLLLSDTNPHPLIHK 206
>TC17275
Length = 796
Score = 30.0 bits (66), Expect = 2.3
Identities = 37/159 (23%), Positives = 62/159 (38%), Gaps = 3/159 (1%)
Frame = +2
Query: 271 EEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHSKSRGNETD 330
EED E + E K L K + + +LSS+ +K+ N +
Sbjct: 164 EEDTENEQEF----EQPLKKKPTKPITTNLSSKNQTKSIKSNNNNLSSKNQTKTTLNAST 331
Query: 331 EVLLEKSGGKIEERNTTT---FTESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRN 387
L + KI++ N +T FT S K KQ + L I + KA
Sbjct: 332 FKKLNSTSIKIKKLNNSTSKSFTNSSK-TSTTSKKQLDLTKLAGAIAKNKTTKATTATDT 508
Query: 388 INSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLD 426
N + + + E EK LN+Q++++++ WM +
Sbjct: 509 KNKIKV----NPGHIEEEK--LNKQSNKKKQSPPNWMFE 607
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 29.3 bits (64), Expect = 3.9
Identities = 18/57 (31%), Positives = 26/57 (45%)
Frame = -2
Query: 41 GNKSQRVMTRRLSLKPVRISAKKPSLHKATCSSTIKDSHFPNHIDLPQEGSSSQGVS 97
G S V R LS PV S + + HK+ ++ +H+DLP S+Q S
Sbjct: 183 GADSTLVPKRFLSSDPVAWSHRSQTTHKSPTQPVACQANAKSHLDLPHASVSTQQFS 13
>TC12615 homologue to AAR23711 (AAR23711) At1g55090, partial (7%)
Length = 555
Score = 29.3 bits (64), Expect = 3.9
Identities = 20/60 (33%), Positives = 31/60 (51%)
Frame = -1
Query: 825 LQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELCKPIKPVETISSCHEEAPT 884
LQS A+ L + LL +V+F+ + T+ + A+ P+P P TISSC + T
Sbjct: 348 LQSSHANL*LISFVLLREVNFNYNPTLGLPEPAAATPIPP--TPSDAATTISSCSSDC*T 175
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 28.9 bits (63), Expect = 5.1
Identities = 29/119 (24%), Positives = 52/119 (43%), Gaps = 13/119 (10%)
Frame = +2
Query: 264 EKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIE------LVQKAFDEILLPEVEDLS 317
++E+EGG E G + + D D DDE++ E LV++ + E E+ S
Sbjct: 122 DEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEEEEEEDLGTEYLVRRT---VAAAEDEEAS 292
Query: 318 SEGH-SKSRGNETDEVLLEKSGGKIEERNT------TTFTESPKEVPKMESKQKSWSHL 369
S+ + G++ D EK+G + + + ++ E + SK+ W HL
Sbjct: 293 SDFEPEEDEGDDNDNDDGEKAGVPSKRKRSDKDGSGDDDSDDGGEDDERPSKR*GWIHL 469
>AI967495
Length = 390
Score = 28.9 bits (63), Expect = 5.1
Identities = 14/39 (35%), Positives = 19/39 (47%)
Frame = +1
Query: 1106 EERKEESVSKEVNKPNQKLSRNWSNLKKVVLLRRFIKAL 1144
E R + VNK N L R W +VV+L+ F+ L
Sbjct: 211 EIRAAPDLKPVVNKHNTSLGRPWQQYSRVVVLKTFMDTL 327
>AV421105
Length = 415
Score = 28.5 bits (62), Expect = 6.7
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Frame = +2
Query: 351 ESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEK-VLL 409
E+PK + ME+K ++W + K K+ +K+ NS P++ PS A++ V
Sbjct: 182 ETPKVMVDMEAKLRAW------LESKGKAKSSQKLGAFNSPLPKKTPSSIITSAKRSVTF 343
Query: 410 NRQTSEERKKSEE 422
N E R + +
Sbjct: 344 NSSVKEGRTATNQ 382
>TC14751 similar to UP|Q948T9 (Q948T9) Respiratory burst oxidase homolog,
partial (34%)
Length = 1160
Score = 28.1 bits (61), Expect = 8.7
Identities = 12/41 (29%), Positives = 20/41 (48%)
Frame = +3
Query: 559 EDLKHGTSTTDVPYGVQERDQKYIKKWHLMYKQAVLSNTGK 599
+ L+H S D+ G + + KW ++K A L + GK
Sbjct: 690 QSLRHAKSGVDIVSGTRVKTHFAKPKWRTVFKPAALKHPGK 812
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.308 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,615,616
Number of Sequences: 28460
Number of extensions: 202437
Number of successful extensions: 1008
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 950
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1005
length of query: 1222
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1121
effective length of database: 2,023,140
effective search space: 2267939940
effective search space used: 2267939940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC125478.8


