

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.9 - phase: 0 /pseudo
(932 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
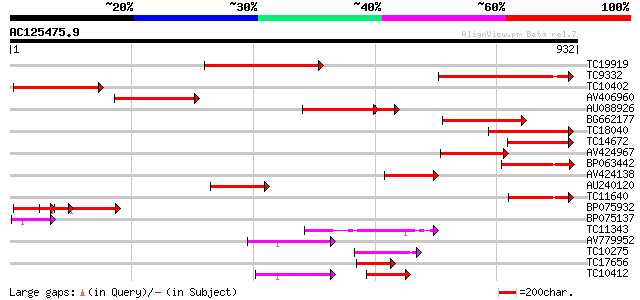
Score E
Sequences producing significant alignments: (bits) Value
TC19919 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPas... 337 5e-93
TC9332 similar to UP|Q9ARG5 (Q9ARG5) Plasma membrane H+ ATPase, ... 302 1e-82
TC10402 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPas... 271 3e-73
AV406960 248 3e-66
AU088926 204 3e-63
BG662177 229 2e-60
TC18040 similar to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPase,... 217 6e-57
TC14672 homologue to UP|Q43106 (Q43106) H(+)-transporting ATPase... 197 5e-51
AV424967 179 2e-45
BP063442 167 5e-42
AV424138 165 3e-41
AU240120 160 1e-39
TC11640 homologue to UP|Q7Y065 (Q7Y065) Plasma membrane H+-ATPas... 147 6e-36
BP075932 137 8e-33
BP075137 86 2e-17
TC11343 similar to PIR|B96660|B96660 protein F2K11.18 [imported]... 78 6e-15
AV779952 58 8e-09
TC10275 similar to UP|Q7Y051 (Q7Y051) Paa2 P-type ATPase, partia... 57 2e-08
TC17656 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transport... 56 2e-08
TC10412 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATP... 56 3e-08
>TC19919 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPase, partial
(21%)
Length = 586
Score = 337 bits (864), Expect = 5e-93
Identities = 164/195 (84%), Positives = 181/195 (92%)
Frame = +1
Query: 321 AIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQDAID 380
AIEEMAGMDVLCSDKTGTLTLNKLSVD+NLIEVF KG++KEHV+LLAARASR ENQDAID
Sbjct: 1 AIEEMAGMDVLCSDKTGTLTLNKLSVDRNLIEVFTKGIEKEHVILLAARASRTENQDAID 180
Query: 381 AAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASKGAPEQIMDLCKL 440
AAIVG LADPKEARAGVRE+HFLPFNPVDKRTALTYID +G+WHRASKGAPEQI++LC
Sbjct: 181 AAIVGMLADPKEARAGVREVHFLPFNPVDKRTALTYIDSDGSWHRASKGAPEQILNLCNC 360
Query: 441 REDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLLSLFDPPRHDSAE 500
+ED ++ H+ IDKFAERGLRSL VARQ VPEK+K++PGAPWQFVGLL LFDPPRHDSAE
Sbjct: 361 KEDVRKRAHSTIDKFAERGLRSLGVARQVVPEKSKDAPGAPWQFVGLLPLFDPPRHDSAE 540
Query: 501 TIRRALHLGVNVKMI 515
TI RAL+LGVNVKMI
Sbjct: 541 TITRALNLGVNVKMI 585
>TC9332 similar to UP|Q9ARG5 (Q9ARG5) Plasma membrane H+ ATPase, partial
(25%)
Length = 960
Score = 302 bits (774), Expect = 1e-82
Identities = 148/223 (66%), Positives = 179/223 (79%), Gaps = 2/223 (0%)
Frame = +1
Query: 706 PDSWKLKEIFATGIVLGGYLALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQ 763
PDSWKLKEIFATG+VLG Y+A+MT +FF+A+ + D F FGV + + D++ SALYLQ
Sbjct: 10 PDSWKLKEIFATGVVLGTYMAIMTAVFFYAVHDTDLFTRLFGVHSIAESEDQLNSALYLQ 189
Query: 764 VSIVSQALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWA 823
VSI+SQALIFVTRSR WSF+ERPG +LV AF AQL+AT+IAVYA+W FA++ GIGW WA
Sbjct: 190 VSIISQALIFVTRSRSWSFVERPGLMLVGAFIAAQLVATVIAVYAHWDFARINGIGWRWA 369
Query: 824 GVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQR 883
GVIW+Y ++ YIPLD++KF IR LSG AW+N+L NKTAFTTKKDYG+ EREAQWA AQR
Sbjct: 370 GVIWIYIVITYIPLDILKFFIRMGLSGSAWDNMLQNKTAFTTKKDYGRGEREAQWAVAQR 549
Query: 884 TLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
TLHGLQ PE+ K+++ E SEIAEQAKRRAE ARL H
Sbjct: 550 TLHGLQVPEA-----HKNNHHEHSEIAEQAKRRAEAARLKELH 663
>TC10402 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPase, partial
(16%)
Length = 613
Score = 271 bits (694), Expect = 3e-73
Identities = 131/148 (88%), Positives = 144/148 (96%)
Frame = +3
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
ISLE+IKNE VDLERIPVEEVFEQLKC+KEGLSS+EGANRLQ+FGPNKLEEK++SK+LKF
Sbjct: 168 ISLEEIKNENVDLERIPVEEVFEQLKCSKEGLSSDEGANRLQVFGPNKLEEKRESKLLKF 347
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANG G+PPDWQDFVGII LL+INSTISFIEENNAGNAA
Sbjct: 348 LGFMWNPLSWVMEAAAIMAIALANGGGRPPDWQDFVGIITLLIINSTISFIEENNAGNAA 527
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILV 154
AALMAGLAPKTKVLRDG+W+EQ+AAILV
Sbjct: 528 AALMAGLAPKTKVLRDGRWTEQDAAILV 611
>AV406960
Length = 425
Score = 248 bits (633), Expect = 3e-66
Identities = 122/139 (87%), Positives = 132/139 (94%)
Frame = +2
Query: 173 LLEGDPLKIDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLV 232
LL+ D L +DQSALTGESLP T++P DEV+SGST K+GE+EAVVIATGVHTFFGKAAHLV
Sbjct: 8 LLDADALSVDQSALTGESLPATKHPSDEVFSGSTVKKGEIEAVVIATGVHTFFGKAAHLV 187
Query: 233 DSTNQVGHFQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPI 292
DSTNQVGHFQKVLTAIGNFCICSIAVG++ E+IVMYPIQHRKYRDGIDNLLVLLIGGIPI
Sbjct: 188 DSTNQVGHFQKVLTAIGNFCICSIAVGIVIELIVMYPIQHRKYRDGIDNLLVLLIGGIPI 367
Query: 293 AMPTVLSVTMAIGSHRLSQ 311
AMPTVLSVTMAIGSHRLSQ
Sbjct: 368 AMPTVLSVTMAIGSHRLSQ 424
>AU088926
Length = 487
Score = 204 bits (519), Expect(2) = 3e-63
Identities = 98/125 (78%), Positives = 110/125 (87%)
Frame = +2
Query: 482 WQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSAT 541
W+F+GLL LFDPPRHDSA TIRRAL LGVNVKMITGDQLAI KETGRRLGMGTNM PS++
Sbjct: 11 WEFLGLLPLFDPPRHDSAXTIRRALDLGVNVKMITGDQLAIGKETGRRLGMGTNMXPSSS 190
Query: 542 LLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPAL 601
LLGQ KDA IA++P++ELIEKADGFAGVFPEHK EIV +L + KHICGMTG GVNDAPAL
Sbjct: 191 LLGQSKDAAIASIPIDELIEKADGFAGVFPEHKXEIVXRLXDTKHICGMTGXGVNDAPAL 370
Query: 602 KRADI 606
+ +
Sbjct: 371 XKXTL 385
Score = 55.8 bits (133), Expect(2) = 3e-63
Identities = 30/40 (75%), Positives = 32/40 (80%)
Frame = +1
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTS 640
++ A IGIAVADATDAA AS IV TEPGL VII AVLTS
Sbjct: 367 IEXAXIGIAVADATDAAXSASHIVXTEPGLXVIIXAVLTS 486
>BG662177
Length = 431
Score = 229 bits (583), Expect = 2e-60
Identities = 110/140 (78%), Positives = 126/140 (89%), Gaps = 2/140 (1%)
Frame = +3
Query: 712 KEIFATGIVLGGYLALMTVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQ 769
+EIFATGIVLGGYLALMTVIFFW MKE FF DKFGVR L+ DEM++ALYLQVSIVSQ
Sbjct: 12 EEIFATGIVLGGYLALMTVIFFWLMKETTFFSDKFGVRPLHESPDEMIAALYLQVSIVSQ 191
Query: 770 ALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLY 829
ALIFVTRSR WS++ERPG LL+ AF IAQLIAT IAVYANWGFA+++G+GWGWA VIW+Y
Sbjct: 192 ALIFVTRSRSWSYVERPGMLLMGAFLIAQLIATFIAVYANWGFARIKGVGWGWARVIWIY 371
Query: 830 SIVFYIPLDVMKFAIRYILS 849
S+VFY+PLD++KFAIRYILS
Sbjct: 372 SVVFYVPLDILKFAIRYILS 431
>TC18040 similar to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPase, partial
(17%)
Length = 747
Score = 217 bits (553), Expect = 6e-57
Identities = 100/139 (71%), Positives = 120/139 (85%)
Frame = +2
Query: 788 ALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYI 847
ALL++AF AQL+AT+IAVYANWGFA ++GIGWGWAGVIWLYSIVFYIPLD++KF IRY
Sbjct: 2 ALLMVAFLAAQLVATLIAVYANWGFADMKGIGWGWAGVIWLYSIVFYIPLDILKFIIRYA 181
Query: 848 LSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELS 907
LSGKAW+N+ +N+ AFT+KKDYGK EREAQWA AQRTLHGL PP++ + NEK+ YRELS
Sbjct: 182 LSGKAWSNITENRVAFTSKKDYGKGEREAQWAAAQRTLHGLDPPDTETMMNEKNHYRELS 361
Query: 908 EIAEQAKRRAEVARLIPFH 926
E+A A++RAEVARL H
Sbjct: 362 ELAGPAQQRAEVARLRELH 418
>TC14672 homologue to UP|Q43106 (Q43106) H(+)-transporting ATPase , partial
(14%)
Length = 828
Score = 197 bits (502), Expect = 5e-51
Identities = 93/108 (86%), Positives = 101/108 (93%)
Frame = +1
Query: 819 GWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQW 878
GWGWAGVIWLY+IV YIPLD++KFAIRY LSGKAW+NLL+NKTAFTTKKDYGKEEREAQW
Sbjct: 1 GWGWAGVIWLYTIVTYIPLDLLKFAIRYALSGKAWDNLLENKTAFTTKKDYGKEEREAQW 180
Query: 879 AHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
A AQRTLHGLQPPE+S +FNEK+SYRELSEIAEQAKRRAEVARL H
Sbjct: 181 AAAQRTLHGLQPPETSNVFNEKNSYRELSEIAEQAKRRAEVARLRELH 324
>AV424967
Length = 344
Score = 179 bits (453), Expect = 2e-45
Identities = 83/114 (72%), Positives = 99/114 (86%), Gaps = 2/114 (1%)
Frame = +1
Query: 709 WKLKEIFATGIVLGGYLALMTVIFFWAMKENDFFPDKFGVRKLNHD--EMMSALYLQVSI 766
WKLKEIFATG+VLG Y+ALMTV+FFW +K+ DFFPDKFGVR + + EMM+ALYLQVSI
Sbjct: 1 WKLKEIFATGVVLGSYMALMTVVFFWLIKDTDFFPDKFGVRSIRNSPGEMMAALYLQVSI 180
Query: 767 VSQALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGW 820
+SQALIFVTRSR WSF+ERPG LL+ AF IAQL+AT +AVYANW FA++ G+GW
Sbjct: 181 ISQALIFVTRSRSWSFVERPGLLLLGAFMIAQLVATFLAVYANWSFARINGMGW 342
>BP063442
Length = 445
Score = 167 bits (424), Expect = 5e-42
Identities = 71/120 (59%), Positives = 97/120 (80%)
Frame = -2
Query: 809 NWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKD 868
NW FA ++GIGW WAGV+WLY+++FYIPLD +KF RY LSG+AW+ +++ + AFT KKD
Sbjct: 444 NWSFAAIEGIGWXWAGVVWLYNLIFYIPLDFIKFITRYALSGRAWDLVIEQRIAFTRKKD 265
Query: 869 YGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFHLC 928
+GKEERE +WAHAQRTLHGL PPE+ +FNE+ +Y +++++AE+AKRRAE+ R LC
Sbjct: 264 FGKEERELKWAHAQRTLHGLHPPETK-MFNERKNYTDINQMAEEAKRRAEITRYAFMCLC 88
>AV424138
Length = 270
Score = 165 bits (418), Expect = 3e-41
Identities = 85/89 (95%), Positives = 87/89 (97%)
Frame = +3
Query: 617 ARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDF 676
AR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDF
Sbjct: 3 ARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDF 182
Query: 677 SPFMVLIIAILNDGTIMTISKDRVVPSPL 705
+PFMVLIIAILNDGTIMTISKDRV PSP+
Sbjct: 183 APFMVLIIAILNDGTIMTISKDRVKPSPM 269
>AU240120
Length = 300
Score = 160 bits (404), Expect = 1e-39
Identities = 75/97 (77%), Positives = 87/97 (89%)
Frame = +3
Query: 330 VLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQDAIDAAIVGTLAD 389
VLCSDKTGTLTLNKL+VDK+LIEVF G DK+ ++L AARASR ENQDAIDA+IV L+D
Sbjct: 9 VLCSDKTGTLTLNKLTVDKSLIEVFPSGFDKDSLVLFAARASRTENQDAIDASIVNMLSD 188
Query: 390 PKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
PKEARAG+ E+HFLPFNPVDKRTA+TYID NG+WHR+
Sbjct: 189 PKEARAGITEVHFLPFNPVDKRTAITYIDSNGDWHRS 299
>TC11640 homologue to UP|Q7Y065 (Q7Y065) Plasma membrane H+-ATPase
(Fragment), partial (34%)
Length = 626
Score = 147 bits (372), Expect = 6e-36
Identities = 63/106 (59%), Positives = 88/106 (82%)
Frame = +3
Query: 821 GWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAH 880
GWAGV+WLY+++FYIPLD +KF RY LSG+AW+ +++ + AFT KKD+GKEERE +WAH
Sbjct: 3 GWAGVVWLYNLIFYIPLDFIKFITRYALSGRAWDLVIEQRIAFTRKKDFGKEERELKWAH 182
Query: 881 AQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
AQRTLHGL PPE+ +F+E+ +Y +++++AE+AKRRAE+ RL H
Sbjct: 183 AQRTLHGLHPPETK-MFHERKNYTDINQMAEEAKRRAEITRLRELH 317
>BP075932
Length = 560
Score = 137 bits (345), Expect = 8e-33
Identities = 77/117 (65%), Positives = 87/117 (73%), Gaps = 9/117 (7%)
Frame = -3
Query: 74 LSWVMEAAALMAIGLAN--GNGKPPDWQD-------FVGIICLLVINSTISFIEENNAGN 124
+SWV ++ M G + GN W + +GIICLLVINSTISFIEENNAGN
Sbjct: 378 VSWVHVESSFMGHGNGSHYGNCSCKWWWEAPRLRKILLGIICLLVINSTISFIEENNAGN 199
Query: 125 AAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKI 181
AAAALMAGLAPK KVLRDGKWSEQEAA+LVPG +LGDIVPADARLLEGDP++I
Sbjct: 198 AAAALMAGLAPKPKVLRDGKWSEQEAAVLVPGRHHKHQLGDIVPADARLLEGDPIQI 28
Score = 87.8 bits (216), Expect = 7e-18
Identities = 47/78 (60%), Positives = 55/78 (70%), Gaps = 10/78 (12%)
Frame = -2
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGP-NKLEEKKDSKILK 65
ISLE+IKNETVDLERIP+EEVF+ LKCT EGLSSEEGA+RLQIFGP + K+ +
Sbjct: 556 ISLEEIKNETVDLERIPIEEVFQHLKCTPEGLSSEEGASRLQIFGPDHATRRKRKANCSS 377
Query: 66 FLG---------FMWNPL 74
FLG + W PL
Sbjct: 376 FLGSCGILFHGSWKWQPL 323
Score = 82.0 bits (201), Expect = 4e-16
Identities = 38/57 (66%), Positives = 44/57 (76%)
Frame = -1
Query: 49 IFGPNKLEEKKDSKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGII 105
I+ + EEKK+SK+LKFLGFMWNPLSWVME AA MAI LANG GKPPD F ++
Sbjct: 428 IWARPRYEEKKESKLLKFLGFMWNPLSWVMEMAATMAIALANGGGKPPDCARFCWVL 258
>BP075137
Length = 507
Score = 86.3 bits (212), Expect = 2e-17
Identities = 49/106 (46%), Positives = 58/106 (54%), Gaps = 34/106 (32%)
Frame = +3
Query: 4 SKSISLEQIKNETVDL----------------------------------ERIPVEEVFE 29
+ IS E +KNE VDL E IPV+EV +
Sbjct: 33 ASDISFEDLKNENVDLVSEIKTKKHRL*CHLRFCFIYLEVTICYIDYYVQENIPVDEVCQ 212
Query: 30 QLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLGFMWNPLS 75
QLKCT+EGLSS+EG RLQ+FGPNKLEE +SK+LK LG MWNP S
Sbjct: 213 QLKCTREGLSSDEGEKRLQMFGPNKLEEVTESKLLKCLGCMWNPRS 350
>TC11343 similar to PIR|B96660|B96660 protein F2K11.18 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(21%)
Length = 909
Score = 78.2 bits (191), Expect = 6e-15
Identities = 64/228 (28%), Positives = 100/228 (43%), Gaps = 7/228 (3%)
Frame = +2
Query: 485 VGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG 544
+G+L++ DP + ++ E + + + M+TGD A R+ G
Sbjct: 164 IGVLAVSDPLKPNAREVVSILNSMNIRSIMVTGDNWGTANSIARQAG------------- 304
Query: 545 QDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRA 604
+E +I +A P+ K V++LQ + M GDG+ND+PAL A
Sbjct: 305 -----------IETVIAEAQ------PQTKATKVKELQTSGYTVAMVGDGINDSPALVAA 433
Query: 605 DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMK-------NYTIYAVSI 657
D+GIA+ TD A ASDIVL L II A+ ++ F R++ Y + A+ I
Sbjct: 434 DVGIAIGAGTDIAIEASDIVLMRSNLEDIIIALDLAKKTFSRIRLNYLWALGYNLLAIPI 613
Query: 658 TIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPL 705
I++ F+ +F P++ G M S RVV S L
Sbjct: 614 AAGILYPFI------RFRLHPWIA--------GVAMAASSVRVVCSSL 715
>AV779952
Length = 557
Score = 57.8 bits (138), Expect = 8e-09
Identities = 39/158 (24%), Positives = 71/158 (44%), Gaps = 14/158 (8%)
Frame = +2
Query: 392 EARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASKGAPEQIMDLC-----------KL 440
+ R V+ + PFN K + +G + KGA E ++ C L
Sbjct: 83 KGRQVVKLVKVEPFNSTKKSMGVVLQLPDGGFRAHCKGASEIVLAACDKVVDSNGEVVPL 262
Query: 441 REDTKRNIHAIIDKFAERGLRSLAVARQEVPEK---TKESPGAPWQFVGLLSLFDPPRHD 497
ED+ + I+KFA+ LR+L + ++ ++ P + + +G++ + DP R
Sbjct: 263 DEDSINQVKDTIEKFADDALRTLCLTYMDIQDEF*VGSPIPTSGYTCIGIVGIKDPVRPG 442
Query: 498 SAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTN 535
E++ G+ V+M+TGD + AK R G+ T+
Sbjct: 443 VRESVAICRSAGITVRMVTGDNIDTAKAIARECGILTD 556
>TC10275 similar to UP|Q7Y051 (Q7Y051) Paa2 P-type ATPase, partial (16%)
Length = 724
Score = 56.6 bits (135), Expect = 2e-08
Identities = 35/113 (30%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Frame = +1
Query: 567 AGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADAT--DAARGASDIV 624
A + P+ K E + L+ H M GDG+NDAPAL AD+GIA+ + +AA A+ I+
Sbjct: 13 ASLAPQQKSEFISSLKAAGHHVAMVGDGINDAPALAVADVGIALQNEAQENAASDAASII 192
Query: 625 LTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDFS 677
L +S ++ A+ +++ ++ +AV+ + I L+ FDF+
Sbjct: 193 LLGNKISQVVDAIDLAQSTMAKVYQNLSWAVAYNV-IAIPIAAGVLLPHFDFA 348
>TC17656 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transporting
ATPase-like protein {Arabidopsis thaliana;} , partial
(25%)
Length = 821
Score = 56.2 bits (134), Expect = 2e-08
Identities = 26/65 (40%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Frame = +3
Query: 571 PEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADA-TDAARGASDIVLTEPG 629
P K +V+ L+ + H+ +TGDG NDAPAL ADIG+A+ A T+ A+ +SDI++ +
Sbjct: 78 PNDKLLLVQALRRKGHVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESSDIIILDDN 257
Query: 630 LSVII 634
+ ++
Sbjct: 258 FASVV 272
>TC10412 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase,
partial (33%)
Length = 1061
Score = 55.8 bits (133), Expect = 3e-08
Identities = 38/146 (26%), Positives = 66/146 (45%), Gaps = 14/146 (9%)
Frame = +2
Query: 404 PFNPVDKRTALTYIDGNGNWHRASKGAPEQIMDLC-----------KLREDTKRNIHAII 452
PFN KR ++ G KGA E ++ C L E++ ++++ I
Sbjct: 47 PFNSTKKRMSVAVELPGGGLRAHCKGASEIVLAACDKVLNSNGEVVPLDEESINHLNSTI 226
Query: 453 DKFAERGLRSLAVARQEVPEKTKESPGAP---WQFVGLLSLFDPPRHDSAETIRRALHLG 509
++FA LR+L +A E+ P + +G++ + DP R E++ G
Sbjct: 227 NQFASEALRTLCLAYMELENGFSAEDPIPLSGFTCIGVVGIKDPVRPGVKESVAVCRSAG 406
Query: 510 VNVKMITGDQLAIAKETGRRLGMGTN 535
+ V+M+TGD + AK R G+ T+
Sbjct: 407 ITVRMVTGDNINTAKAIARECGILTD 484
Score = 53.9 bits (128), Expect = 1e-07
Identities = 22/74 (29%), Positives = 47/74 (62%), Gaps = 1/74 (1%)
Frame = +3
Query: 587 ICGMTGDGVNDAPALKRADIGIAVADA-TDAARGASDIVLTEPGLSVIISAVLTSRAIFQ 645
+ +TGDG NDAPAL ADIG+A+ A T+ A+ ++D+++ + S I++ R+++
Sbjct: 654 VVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYI 833
Query: 646 RMKNYTIYAVSITI 659
++ + + +++ +
Sbjct: 834 NIQKFVQFQLTVNV 875
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,937,215
Number of Sequences: 28460
Number of extensions: 169495
Number of successful extensions: 901
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 886
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 894
length of query: 932
length of database: 4,897,600
effective HSP length: 99
effective length of query: 833
effective length of database: 2,080,060
effective search space: 1732689980
effective search space used: 1732689980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC125475.9


