

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.5 + phase: 0 /pseudo
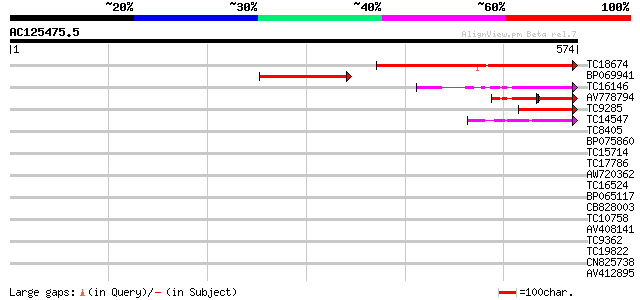
(574 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18674 similar to UP|Q7Y0V3 (Q7Y0V3) Actin filament bundling pr... 283 5e-77
BP069941 111 3e-25
TC16146 weakly similar to UP|VIL4_ARATH (O65570) Villin 4, parti... 96 1e-20
AV778794 54 3e-13
TC9285 weakly similar to PIR|T50669|T50669 villin 2 [imported] -... 67 1e-11
TC14547 weakly similar to UP|VILI_CHICK (P02640) Villin, partial... 61 6e-10
TC8405 weakly similar to UP|O04083 (O04083) Lysophospholipase is... 35 0.043
BP075860 34 0.074
TC15714 weakly similar to UP|Q9LU96 (Q9LU96) Genomic DNA, chromo... 33 0.13
TC17786 similar to PIR|T46177|T46177 villin 3 homolog T8H10.10 -... 32 0.21
AW720362 32 0.37
TC16524 similar to GB|AAL69537.1|18491137|AY074839 AT5g45420/MFC... 31 0.48
BP065117 31 0.62
CB828003 31 0.62
TC10758 30 1.1
AV408141 30 1.1
TC9362 similar to PIR|F86340|F86340 protein F2D10.34 [imported] ... 30 1.4
TC19822 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 prec... 29 1.8
CN825738 28 3.1
AV412895 28 3.1
>TC18674 similar to UP|Q7Y0V3 (Q7Y0V3) Actin filament bundling protein
P-115-ABP, partial (17%)
Length = 952
Score = 283 bits (724), Expect = 5e-77
Identities = 148/208 (71%), Positives = 168/208 (80%), Gaps = 5/208 (2%)
Frame = +2
Query: 372 SYGGRSGGLPEKSQRS-RSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRK 430
+YGGRS +P+KSQRS RSMSVSPDRVRVRGRSPAFNALAA FEN RNLSTPPP++RK
Sbjct: 2 AYGGRSSSVPDKSQRSSRSMSVSPDRVRVRGRSPAFNALAANFENPGARNLSTPPPVVRK 181
Query: 431 LYPKSKTPDLATLAPKSSAISHLTSTFE-PPSAREKLIPRSLK---DTSKSNPETNSDNE 486
LYPKSKTPD A LAPKS+AI+ LTS+FE PPSARE +IPRS+K T KSNP+ N D E
Sbjct: 182 LYPKSKTPDSAILAPKSAAIAALTSSFEQPPSARETMIPRSVKVSPVTPKSNPDKN-DKE 358
Query: 487 NSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEF 546
NS SR ESLTI+EDV EGE ED EGL +PYE +K S DP+P IDVTKRE YLS EF
Sbjct: 359 NSASSRVESLTIEEDVKEGEAEDEEGLSFHPYERLKVSSPDPVPGIDVTKRETYLSSAEF 538
Query: 547 QERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+E+ GM++ FYKLPKWKQN LKMA+QL
Sbjct: 539 KEKFGMSKDAFYKLPKWKQNXLKMAIQL 622
>BP069941
Length = 304
Score = 111 bits (278), Expect = 3e-25
Identities = 49/93 (52%), Positives = 68/93 (72%)
Frame = +1
Query: 254 EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISC 313
EE++NFSQDDL+TEDI ILD H+++FVW+GQ VD K + A I +K++++ LE +S
Sbjct: 25 EEVYNFSQDDLLTEDILILDTHAEVFVWIGQSVDTKEKQNAFEIAQKYIDKATSLEGLSP 204
Query: 314 SAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNS 346
P+Y V EG+EP FFT +F WD AK+ + GNS
Sbjct: 205 HVPLYKVTEGNEPCFFTTYFSWDHAKATVQGNS 303
>TC16146 weakly similar to UP|VIL4_ARATH (O65570) Villin 4, partial (6%)
Length = 689
Score = 96.3 bits (238), Expect = 1e-20
Identities = 62/162 (38%), Positives = 87/162 (53%)
Frame = +2
Query: 413 FENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLK 472
F S R LS+P P+++KL+ S S+ E+ +P++
Sbjct: 11 FRQSGDRLLSSPSPVVKKLFDGSPAN----------------------SSAEQTLPQA-- 118
Query: 473 DTSKSNPETNSDNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDI 532
+P T E S+ + ES T Q+D N D E L VYPYE ++ S +P+ I
Sbjct: 119 ----DSPAT----ELSSSNERESFT-QKDRNV----DGENLLVYPYEHLRVVSANPVTGI 259
Query: 533 DVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
D+T+RE YLS EEF E+ GM +S FYKLP+WKQNKLKM++ L
Sbjct: 260 DITRRETYLSNEEFHEKFGMPKSAFYKLPRWKQNKLKMSLDL 385
>AV778794
Length = 545
Score = 53.5 bits (127), Expect(2) = 3e-13
Identities = 23/39 (58%), Positives = 30/39 (75%)
Frame = -2
Query: 536 KREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
K YLS EEF E+ GM +S FYKLP+WK+N+LKM++ L
Sbjct: 334 K*HTYLSNEEFHEKFGMPKSAFYKLPRWKRNQLKMSLDL 218
Score = 38.5 bits (88), Expect(2) = 3e-13
Identities = 22/51 (43%), Positives = 31/51 (60%)
Frame = -1
Query: 488 STGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKRE 538
S+ + ES T Q+D N D E L VYPYE ++ S +P+ ID+T+RE
Sbjct: 542 SSSNERESFT-QKDRNV----DGENLLVYPYEHLRVVSANPVTGIDITRRE 405
>TC9285 weakly similar to PIR|T50669|T50669 villin 2 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(7%)
Length = 657
Score = 66.6 bits (161), Expect = 1e-11
Identities = 30/59 (50%), Positives = 41/59 (68%)
Frame = +1
Query: 516 YPYESVKTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+ Y+ +KT S +P ID+ +REAYLS EEF+ +GM + FYKLPKWKQ+ LK +L
Sbjct: 34 FSYDQLKTTSGKNVPGIDLKRREAYLSDEEFETVIGMVKDAFYKLPKWKQDLLKKKFEL 210
>TC14547 weakly similar to UP|VILI_CHICK (P02640) Villin, partial (5%)
Length = 794
Score = 60.8 bits (146), Expect = 6e-10
Identities = 39/111 (35%), Positives = 57/111 (51%)
Frame = +2
Query: 464 EKLIPRSLKDTSKSNPETNSDNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKT 523
E+ + K+ + PET GS +S QE+V E E + + YE +K
Sbjct: 35 EEALEEEXKEREELAPET--------GSNGDSEPKQENV-EDENDSQNSQRFFSYEKLKA 187
Query: 524 DSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
S + +D+ +REAYLS +EF+ GMT+ F KLP+WK + LK V L
Sbjct: 188 KSE--LSGVDLKRREAYLSGKEFETVFGMTKEAFSKLPRWKPDMLKRKVDL 334
>TC8405 weakly similar to UP|O04083 (O04083) Lysophospholipase isolog,
25331-24357 (At1g11090), partial (18%)
Length = 552
Score = 34.7 bits (78), Expect = 0.043
Identities = 35/115 (30%), Positives = 47/115 (40%), Gaps = 8/115 (6%)
Frame = +3
Query: 386 RSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAP 445
RS SP + + S A A T S+ R+ ST PP P S AT AP
Sbjct: 156 RSSPSPSSPSTPKSKPPSS*PTATAPTPAGSSRRSASTSPPGAT---PSSPPTSSATAAP 326
Query: 446 KSSAISHLTSTFEPP--------SAREKLIPRSLKDTSKSNPETNSDNENSTGSR 492
+SA + TST PP SA P + +S ++P S + T +R
Sbjct: 327 TASAATSATSTASPPPRSPSSSTSAAASPTP-PFRPSSSASPWAGSPPSSCTSNR 488
>BP075860
Length = 403
Score = 33.9 bits (76), Expect = 0.074
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Frame = -2
Query: 520 SVKTDSTDPMPDIDVTKREAYLSPEEFQER-LGMTRSEFYKLPKWKQNKLKMAVQL 574
S +T S +P ID+ +REA LS E L + F KLP WKQ LK +L
Sbjct: 351 SYQTTSG*SVPGIDLKRREASLSDAEL*NL*LEW*KMHFTKLPYWKQTLLKKKFEL 184
>TC15714 weakly similar to UP|Q9LU96 (Q9LU96) Genomic DNA, chromosome 3, P1
clone: MPE11, partial (26%)
Length = 1157
Score = 33.1 bits (74), Expect = 0.13
Identities = 21/61 (34%), Positives = 28/61 (45%)
Frame = +2
Query: 423 TPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETN 482
+PPP P+S+TP L + P SS I + TS PP R S + + S T
Sbjct: 377 SPPPT-----PRSRTPSLISAPPTSSPICYTTSLSLPPPTRSSSRTTSPRSSWLSRAVTR 541
Query: 483 S 483
S
Sbjct: 542 S 544
>TC17786 similar to PIR|T46177|T46177 villin 3 homolog T8H10.10 -
Arabidopsis thaliana (fragment) {Arabidopsis thaliana;},
partial (42%)
Length = 1228
Score = 32.3 bits (72), Expect = 0.21
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Frame = +3
Query: 38 ITSQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHL-IGTWIGKNS 91
+ +++WR+ + L ++ KFY GD YI + G+ I WIGK++
Sbjct: 357 VGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLFDIHFWIGKDT 521
>AW720362
Length = 479
Score = 31.6 bits (70), Expect = 0.37
Identities = 25/96 (26%), Positives = 42/96 (43%)
Frame = +2
Query: 377 SGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSK 436
SG + RS SP R R + A++ ++ R+ P P R + +S
Sbjct: 83 SGPVQRHEDRS*RRGGSP-----RSRCGCYEV*CASYVGTDCRDARNPRPDARPRHCRSL 247
Query: 437 TPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLK 472
++ + AP + A + S PPS+R +PR L+
Sbjct: 248 --NIPSSAPHAPACAKRHSRTSPPSSRRNRLPRPLQ 349
>TC16524 similar to GB|AAL69537.1|18491137|AY074839 AT5g45420/MFC19_9
{Arabidopsis thaliana;}, partial (41%)
Length = 997
Score = 31.2 bits (69), Expect = 0.48
Identities = 28/87 (32%), Positives = 38/87 (43%), Gaps = 4/87 (4%)
Frame = +1
Query: 356 NGGTPPLVKPKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNA----LAA 411
+ +P L P S S S P S RS S S SP SP F++ LA+
Sbjct: 16 SSSSPSLSLPSSSPSHS----SSSNPNPSNRSSSGSPSPSS---SAPSPRFHSPAAMLAS 174
Query: 412 TFENSNVRNLSTPPPMIRKLYPKSKTP 438
E S + ++TPPP + P+S P
Sbjct: 175 AAETS*ISLMNTPPPRKIRSLPRSGDP 255
>BP065117
Length = 546
Score = 30.8 bits (68), Expect = 0.62
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = +3
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREE-HLIGTWIGKNS 91
+++WR+ + L ++ KFY GD YI + G+ + + WIGK++
Sbjct: 177 TEIWRIENFQPVPLPKSEYGKFYMGDSYIILQTRQGKGGTYFYDLHFWIGKDT 335
>CB828003
Length = 442
Score = 30.8 bits (68), Expect = 0.62
Identities = 22/72 (30%), Positives = 29/72 (39%)
Frame = +2
Query: 424 PPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNS 483
PPP R P S P + + K SH +STF P + T SNP ++
Sbjct: 101 PPPATRNSSPAS-APSSSHRSKKPETGSHRSSTFSAP-----------RSTRTSNPLQST 244
Query: 484 DNENSTGSREES 495
N+ SR S
Sbjct: 245 PTPNAVASRSSS 280
>TC10758
Length = 532
Score = 30.0 bits (66), Expect = 1.1
Identities = 20/73 (27%), Positives = 28/73 (37%)
Frame = +2
Query: 391 SVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAI 450
S SP ++ R +P+ +N + S PPP + Y P T P S
Sbjct: 164 SPSPQKISARSSNPS--------PRTNSWSFSNPPPSVTPTYSTPSAPS-PTATPPSENS 316
Query: 451 SHLTSTFEPPSAR 463
S S +PP R
Sbjct: 317 SSAASPAKPPPRR 355
>AV408141
Length = 427
Score = 30.0 bits (66), Expect = 1.1
Identities = 34/119 (28%), Positives = 51/119 (42%), Gaps = 10/119 (8%)
Frame = +2
Query: 362 LVKPKRR--ASVSYGGRSGGLPEKSQRSRSMS----VSPDRVRVRGRSPAFNALAA--TF 413
L P RR S + ++ +P S R+ S + VS + R P L++ T
Sbjct: 41 LTDPPRRNPMSTTSARKTSVMPTSSPRAPSPTPSAPVSVPKAWTR*FPPPPTRLSSPTTV 220
Query: 414 ENSNVR-NLSTPPPMIRKLYPKSKTPDLATLAPKSS-AISHLTSTFEPPSAREKLIPRS 470
S+ R S PPP +P +TP AT P SS +++H +S+ S P S
Sbjct: 221 PPSSTRCRSSNPPPRCSSSFPSLRTPQPATEPPPSSLSLAHSSSSASSSSPAASTPPSS 397
>TC9362 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(58%)
Length = 591
Score = 29.6 bits (65), Expect = 1.4
Identities = 15/42 (35%), Positives = 23/42 (54%)
Frame = +2
Query: 421 LSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSA 462
+STP P + P TP ++ +P SSA+S +S + P A
Sbjct: 47 VSTPRPPPHLIPPPPSTPTSSSSSPLSSALSSASSAWSPSPA 172
>TC19822 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precursor
(At-EXP13) (AtEx13) (Ath-ExpAlpha-1.22), partial (27%)
Length = 532
Score = 29.3 bits (64), Expect = 1.8
Identities = 23/73 (31%), Positives = 30/73 (40%), Gaps = 1/73 (1%)
Frame = -2
Query: 419 RNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAR-EKLIPRSLKDTSKS 477
RN PPP P + P L T +A+ +T+ P SAR + RS S S
Sbjct: 381 RNNHPPPPNGSPPPPHTTPPQLLTQWTARAAMETVTARPPPHSARLSSVAARSAARASSS 202
Query: 478 NPETNSDNENSTG 490
S + STG
Sbjct: 201 GALRRSQRQRSTG 163
>CN825738
Length = 335
Score = 28.5 bits (62), Expect = 3.1
Identities = 26/79 (32%), Positives = 35/79 (43%)
Frame = -1
Query: 424 PPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNS 483
PP + L P S P L SS+ S + F P S + + L TS E+ S
Sbjct: 335 PPSLTSFLIPSSTKP----LPLTSSSASVIP--FSPTSTKSTIPFSPLPPTSSKGAESIS 174
Query: 484 DNENSTGSREESLTIQEDV 502
+ NS+ S SLT +E V
Sbjct: 173 FSGNSSKSLFSSLTREESV 117
>AV412895
Length = 405
Score = 28.5 bits (62), Expect = 3.1
Identities = 12/23 (52%), Positives = 13/23 (56%)
Frame = +3
Query: 323 GSEPPFFTRFFKWDSAKSAMLGN 345
GSE PF T FF W+ S GN
Sbjct: 324 GSESPFITVFFNWEK*SSEARGN 392
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,007,086
Number of Sequences: 28460
Number of extensions: 136683
Number of successful extensions: 925
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 908
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 921
length of query: 574
length of database: 4,897,600
effective HSP length: 95
effective length of query: 479
effective length of database: 2,193,900
effective search space: 1050878100
effective search space used: 1050878100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC125475.5


