

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124972.9 + phase: 0 /pseudo
(527 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
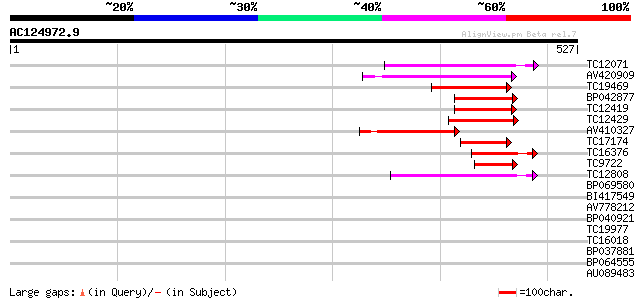
Score E
Sequences producing significant alignments: (bits) Value
TC12071 weakly similar to GB|AAP31968.1|30387605|BT006624 At2g04... 97 6e-21
AV420909 97 8e-21
TC19469 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, par... 96 2e-20
BP042877 88 4e-18
TC12419 81 5e-16
TC12429 similar to UP|Q9FKQ1 (Q9FKQ1) Emb|CAB89401.1 (AT5g65380/... 80 8e-16
AV410327 74 5e-14
TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120,... 71 5e-13
TC16376 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (17%) 66 1e-11
TC9722 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (16%) 55 3e-08
TC12808 weakly similar to GB|CAA16564.1|2827556|ATT12H17 predict... 51 4e-07
BP069580 40 0.001
BI417549 40 0.001
AV778212 34 0.052
BP040921 32 0.33
TC19977 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (20%) 31 0.44
TC16018 UP|Q8MZB8 (Q8MZB8) AT15479p (CG31542-PA), partial (7%) 31 0.57
BP037881 29 1.7
BP064555 28 2.8
AU089483 28 4.8
>TC12071 weakly similar to GB|AAP31968.1|30387605|BT006624 At2g04100
{Arabidopsis thaliana;}, partial (32%)
Length = 764
Score = 97.1 bits (240), Expect = 6e-21
Identities = 56/143 (39%), Positives = 84/143 (58%)
Frame = +3
Query: 349 GLGAAASVRVSNELGAAHPRVAKFSVFVVNGNSMLISVIFAAIILILRVGLSRLFTSDS* 408
G+GAA S RVSNELGA P+ A+ ++F V + L ++I ++ + R L F++D
Sbjct: 27 GVGAAVSTRVSNELGAGKPKEARDAIFAVIILATLDAIILSSALFCCRHVLGFAFSNDLE 206
Query: 409 VIEAVSDLTPLLAISVLLNGIQPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFK 468
V+ +V+ + PLL +SV ++ +LSGVA G GWQ + A NL+ YY IG+ V +LGF
Sbjct: 207 VVHSVAKIVPLLCLSVCVDSFLGVLSGVARGAGWQKIGAVTNLLAYYAIGIPVALLLGFV 386
Query: 469 TSLGVARSTMNLIVIYIGHLVGN 491
L N ++IG L G+
Sbjct: 387 LKL-------NAKGLWIGILTGS 434
>AV420909
Length = 421
Score = 96.7 bits (239), Expect = 8e-21
Identities = 57/143 (39%), Positives = 87/143 (59%)
Frame = +3
Query: 329 SKCTSIFSMNYLNWDMQVMLGLGAAASVRVSNELGAAHPRVAKFSVFVVNGNSMLISVIF 388
S C + +++Y V +GAAAS RVSNELGA +P+ AK +V VV ++ ++I
Sbjct: 3 SVCLNTTTLHYF-----VTYAVGAAASTRVSNELGAGNPKTAKGAVRVVVIVGIVEAIIV 167
Query: 389 AAIILILRVGLSRLFTSDS*VIEAVSDLTPLLAISVLLNGIQPILSGVAIGCGWQALVAY 448
+A + R L +++D V++ V+D+ PLL SV+ + + LSGVA G G+Q + AY
Sbjct: 168 SAFFICFRNVLGYAYSNDKEVVDYVADMAPLLCGSVIGDSLIGALSGVARGGGFQQIGAY 347
Query: 449 VNLVCYYVIGLTVGCVLGFKTSL 471
NL YYV+G+ +G +LGF L
Sbjct: 348 ANLGAYYVVGIPIGLLLGFHLHL 416
>TC19469 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (26%)
Length = 501
Score = 95.5 bits (236), Expect = 2e-20
Identities = 45/74 (60%), Positives = 59/74 (78%)
Frame = +1
Query: 393 LILRVGLSRLFTSDS*VIEAVSDLTPLLAISVLLNGIQPILSGVAIGCGWQALVAYVNLV 452
L LR L+ +FT DS V EAV DL+PLL+IS+LLN + P+LSGV++G GW ++VAYVN+
Sbjct: 1 LFLRERLAYVFTLDSEVAEAVGDLSPLLSISILLNSVPPVLSGVSVGAGWPSVVAYVNIG 180
Query: 453 CYYVIGLTVGCVLG 466
CYY+IG+ VG VLG
Sbjct: 181 CYYLIGIPVGVVLG 222
>BP042877
Length = 489
Score = 87.8 bits (216), Expect = 4e-18
Identities = 39/59 (66%), Positives = 48/59 (81%)
Frame = -1
Query: 414 SDLTPLLAISVLLNGIQPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFKTSLG 472
SDL PLLA+S+LL+GIQP+LSGVA+GCGW A VAYVN+ CYY +G+ +G VLGF G
Sbjct: 489 SDLCPLLALSILLHGIQPVLSGVAVGCGWPAFVAYVNVGCYYGVGIPLGSVLGFYFQFG 313
>TC12419
Length = 667
Score = 80.9 bits (198), Expect = 5e-16
Identities = 37/58 (63%), Positives = 46/58 (78%)
Frame = +3
Query: 414 SDLTPLLAISVLLNGIQPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFKTSL 471
+ + PLLAIS+LL+ IQ +LSGVA GCGWQ L AYVNL +Y+IGL + C+L FKTSL
Sbjct: 9 ASVAPLLAISILLDSIQGVLSGVARGCGWQHLAAYVNLATFYLIGLPISCLLAFKTSL 182
>TC12429 similar to UP|Q9FKQ1 (Q9FKQ1) Emb|CAB89401.1 (AT5g65380/MNA5_11),
partial (22%)
Length = 523
Score = 80.1 bits (196), Expect = 8e-16
Identities = 34/65 (52%), Positives = 53/65 (81%)
Frame = +1
Query: 409 VIEAVSDLTPLLAISVLLNGIQPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFK 468
V++ V++L+ LLA ++LLN IQP+LSGVA+G GWQ+ VAY+NL CYY++G+ +G ++G+
Sbjct: 1 VLDEVNNLSLLLAFTILLNSIQPVLSGVAVGSGWQSYVAYINLGCYYLVGVPLGFIMGWV 180
Query: 469 TSLGV 473
+ GV
Sbjct: 181 FNQGV 195
>AV410327
Length = 424
Score = 74.3 bits (181), Expect = 5e-14
Identities = 46/93 (49%), Positives = 57/93 (60%)
Frame = +2
Query: 326 DSPSKCTSIFSMNYLNWDMQVMLGLGAAASVRVSNELGAAHPRVAKFSVFVVNGNSMLIS 385
DS S CT+I W + +G AAASVRVSNELGA +P+ A FSV VV S ++S
Sbjct: 161 DSLSICTTISG-----WVFMISVGFNAAASVRVSNELGARNPKSASFSVVVVTLISFIMS 325
Query: 386 VIFAAIILILRVGLSRLFTSDS*VIEAVSDLTP 418
VI A ++L LR +S +FT V AVSDL P
Sbjct: 326 VIAALVVLALRDIISYVFTGGEEVAAAVSDLCP 424
>TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120, partial
(18%)
Length = 723
Score = 70.9 bits (172), Expect = 5e-13
Identities = 31/47 (65%), Positives = 39/47 (82%)
Frame = +1
Query: 420 LAISVLLNGIQPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLG 466
LA+S+LL + P+LSGVA+G GW++ VAYVNL CYY+IGL VG VLG
Sbjct: 1 LAVSILLYSVPPVLSGVALGAGWRSTVAYVNLGCYYIIGLPVGIVLG 141
>TC16376 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (17%)
Length = 550
Score = 66.2 bits (160), Expect = 1e-11
Identities = 32/61 (52%), Positives = 39/61 (63%)
Frame = +2
Query: 430 QPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFKTSLGVARSTMNLIVIYIGHLV 489
QP+LSGVA+GCGWQ VAYVN+ CYY IG+ G VLGF G + I++G L
Sbjct: 2 QPVLSGVAVGCGWQTFVAYVNVGCYYGIGIPFGAVLGFYFKFGA-------MGIWLGMLA 160
Query: 490 G 490
G
Sbjct: 161 G 163
>TC9722 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (16%)
Length = 571
Score = 55.1 bits (131), Expect = 3e-08
Identities = 24/40 (60%), Positives = 29/40 (72%)
Frame = +3
Query: 433 LSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFKTSLG 472
LSGVA+GCG Q VAYVN+ CYY +G+ +G VLGF G
Sbjct: 3 LSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFG 122
>TC12808 weakly similar to GB|CAA16564.1|2827556|ATT12H17 predicted protein
{Arabidopsis thaliana;}, partial (32%)
Length = 624
Score = 51.2 bits (121), Expect = 4e-07
Identities = 40/136 (29%), Positives = 61/136 (44%)
Frame = +1
Query: 355 SVRVSNELGAAHPRVAKFSVFVVNGNSMLISVIFAAIILILRVGLSRLFTSDS*VIEAVS 414
S RVSNELGA A S V +++ I + +++ R LF+ D +I V
Sbjct: 4 STRVSNELGANQAGRAYRSACVSLALGLILGCIGSLVMVAARGIWGPLFSHDRGIINGVK 183
Query: 415 DLTPLLAISVLLNGIQPILSGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFKTSLGVA 474
L+A+ + N + G+ G L Y NL +Y + L +G V FK +LG
Sbjct: 184 KTMLLMALVEVFNFPLAVCGGIVRGTARPWLGMYANLGGFYFLALPLGVVFAFKLNLG-- 357
Query: 475 RSTMNLIVIYIGHLVG 490
L ++IG + G
Sbjct: 358 -----LFGLFIGLITG 390
>BP069580
Length = 483
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/39 (43%), Positives = 25/39 (63%)
Frame = -1
Query: 434 SGVAIGCGWQALVAYVNLVCYYVIGLTVGCVLGFKTSLG 472
SG A GCG Q + A++NL YY++G+ + VL F +G
Sbjct: 264 SGTARGCGLQKIGAFINLGSYYLVGIPLAMVLAFVLHIG 148
>BI417549
Length = 404
Score = 39.7 bits (91), Expect = 0.001
Identities = 31/112 (27%), Positives = 53/112 (46%), Gaps = 7/112 (6%)
Frame = +1
Query: 292 GTGFYFHLSISI-------WYVNNFSLNNVSCLFVNMLSNHDSPSKCTSIFSMNYLNWDM 344
G G +F LS+ W+ + L ++ LF N S C +I ++++
Sbjct: 61 GLGEFFRLSVPSAAMVCVKWWASEL-LVLLAGLFSNPELETSILSICLTISTLHFT---- 225
Query: 345 QVMLGLGAAASVRVSNELGAAHPRVAKFSVFVVNGNSMLISVIFAAIILILR 396
+ G G AAS RV+NELGA +P+ A+ V + + + +VI + + R
Sbjct: 226 -IPYGFGVAASTRVANELGAGNPKAARVVVSIAMSLAFIEAVIVSTALFGFR 378
>AV778212
Length = 557
Score = 34.3 bits (77), Expect = 0.052
Identities = 18/45 (40%), Positives = 24/45 (53%)
Frame = +2
Query: 91 LLEFKVLLMALC*EWQVQCKMYVDKLMEPKNMQQCASLCKEHSSY 135
++E LMA C EW+ + K Y DK N +CKEHSS+
Sbjct: 284 MMESNSSLMA*CWEWEARWKHYADKHTVLTNTIC*EFICKEHSSF 418
>BP040921
Length = 520
Score = 31.6 bits (70), Expect = 0.33
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = -1
Query: 168 KCSLMDLFLNSMHLHSAVPCRGFFKHR 194
+ +LM LF +S +HS+ PC G F R
Sbjct: 199 RVTLMSLFTSSFSVHSSTPCHGMFSSR 119
>TC19977 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (20%)
Length = 452
Score = 31.2 bits (69), Expect = 0.44
Identities = 18/34 (52%), Positives = 19/34 (54%)
Frame = +1
Query: 88 P*PLLEFKVLLMALC*EWQVQCKMYVDKLMEPKN 121
P L K LM LC* W VQ K YVDK M K+
Sbjct: 346 PLETLASKSSLMDLC*AWGVQWKHYVDKHMVQKS 447
>TC16018 UP|Q8MZB8 (Q8MZB8) AT15479p (CG31542-PA), partial (7%)
Length = 425
Score = 30.8 bits (68), Expect = 0.57
Identities = 15/36 (41%), Positives = 21/36 (57%), Gaps = 1/36 (2%)
Frame = -2
Query: 18 IANLSSDDVEELLEHRPVELRWWLKL-VAWESWLLW 52
+ ++S DV +EH LRWW + +A E WLLW
Sbjct: 349 MGHVSICDVVRCVEHVGFWLRWWWTVRLAVEGWLLW 242
>BP037881
Length = 362
Score = 29.3 bits (64), Expect = 1.7
Identities = 28/86 (32%), Positives = 35/86 (40%), Gaps = 10/86 (11%)
Frame = +1
Query: 310 SLNNVSCLFVNMLSNHDSPSKCTSIFSMNYLNWDMQVMLGL---------GAAASVRV-S 359
SL N F N+ K S F Y+N D V LGL G VRV
Sbjct: 109 SLKNYIKEFKNLSKTETEQHKGWS*F---YINSDSDVSLGLNIYSFTKFSGETTRVRVLD 279
Query: 360 NELGAAHPRVAKFSVFVVNGNSMLIS 385
N G P ++K ++V+ G LIS
Sbjct: 280 NYHGWVFPELSKHRIYVLKGGVSLIS 357
>BP064555
Length = 523
Score = 28.5 bits (62), Expect = 2.8
Identities = 17/39 (43%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Frame = -2
Query: 293 TGFYFHLSISIWYVNNFSLNNVSCLFVNMLSNHD-SPSK 330
T FYF+LS I Y ++ L +++C F + LS+H SPS+
Sbjct: 477 TSFYFYLSHLIIYHISYFL-SLTCFFSSYLSHHSTSPSQ 364
>AU089483
Length = 396
Score = 27.7 bits (60), Expect = 4.8
Identities = 17/80 (21%), Positives = 34/80 (42%), Gaps = 16/80 (20%)
Frame = -3
Query: 297 FHLSISIWYVNNF------SLNNVSC-----LFVNMLSNHDSPSKCTSIFSMN-----YL 340
F + IW++NN S+ ++ C L + +L P +F N ++
Sbjct: 244 FRXPVCIWFINNSTKI*NDSITHIRCSESQPLIIQVLKPQSPPHLALQVFKYNENFHLFI 65
Query: 341 NWDMQVMLGLGAAASVRVSN 360
N + + L L + S+ +S+
Sbjct: 64 NLSLSLSLSLSLSLSLSLSS 5
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.345 0.150 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,626,682
Number of Sequences: 28460
Number of extensions: 169926
Number of successful extensions: 2112
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 2068
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2106
length of query: 527
length of database: 4,897,600
effective HSP length: 94
effective length of query: 433
effective length of database: 2,222,360
effective search space: 962281880
effective search space used: 962281880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC124972.9


