

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.12 - phase: 0
(552 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
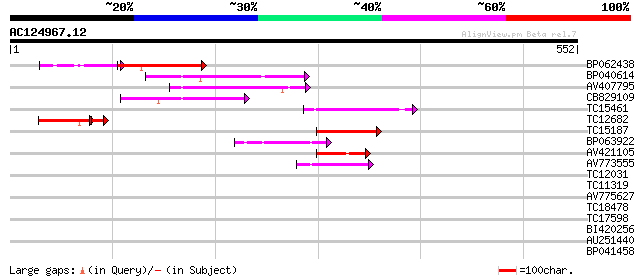
Score E
Sequences producing significant alignments: (bits) Value
BP062438 80 9e-19
BP040614 77 9e-15
AV407795 74 6e-14
CB829109 67 6e-12
TC15461 similar to UP|O24147 (O24147) Kinesin-like protein, part... 61 4e-10
TC12682 52 4e-08
TC15187 similar to UP|O24147 (O24147) Kinesin-like protein, part... 54 5e-08
BP063922 44 7e-05
AV421105 43 1e-04
AV773555 43 2e-04
TC12031 homologue to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-l... 38 0.005
TC11319 31 0.60
AV775627 28 3.0
TC18478 homologue to UP|UAP1_ARATH (O64765) Probable UDP-N-acety... 28 3.0
TC17598 weakly similar to GB|AAP68293.1|31711874|BT008854 At1g80... 28 5.0
BI420256 27 6.6
AU251440 27 8.6
BP041458 27 8.6
>BP062438
Length = 498
Score = 80.1 bits (196), Expect(2) = 9e-19
Identities = 42/92 (45%), Positives = 60/92 (64%), Gaps = 6/92 (6%)
Frame = +3
Query: 106 GINATIFAYGQTSSGKTFTMRG------ITENAIRDIYECIKNTPDRDFVLKISALEIYN 159
G N T+FAYGQT+SGKT TMRG + A+RD+++ I+ DR+F+L++S +EIYN
Sbjct: 222 GFNGTVFAYGQTNSGKTHTMRGSKADPGVIPRAVRDLFQIIQQDVDREFLLRMSYMEIYN 401
Query: 160 ETVIDLLNRESGPLRILDDTEKVTVVENLFEE 191
E + DLL E L+I ++ E+ V L EE
Sbjct: 402 EEINDLLAPEHRKLQIHENLERGIYVAGLREE 497
Score = 30.4 bits (67), Expect(2) = 9e-19
Identities = 24/83 (28%), Positives = 36/83 (42%)
Frame = +2
Query: 30 EKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACSTQ 89
E+I VTVR RPL+ W + + PN S+ + FD++F C T
Sbjct: 20 ERIHVTVRARPLS---PGGAKTSLWRISGNSIYL---PNH---SSKFEFDQIFSDNCETS 172
Query: 90 KVYDEGAKDVALSALSGINATIF 112
+VY K + +A + I F
Sbjct: 173 QVYHARTKHIVEAAGAWIQRYCF 241
>BP040614
Length = 504
Score = 76.6 bits (187), Expect = 9e-15
Identities = 56/165 (33%), Positives = 86/165 (51%), Gaps = 5/165 (3%)
Frame = +1
Query: 133 IRDIYECIKNTPDR-DFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTV----VEN 187
+RD++ K+ D + + + +EIYNE V DLL +G R LD + V +
Sbjct: 22 LRDLFHISKDRADSIKYEVFVQMIEIYNEQVRDLLV-SNGSNRRLDIRNNSQLNGLNVPD 198
Query: 188 LFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYI 247
+ Q + +L+ + + +R VG T LN++SSRSH ++ + V S ++
Sbjct: 199 AYLVPVTCTQDVLYLMEVGQKNRAVGATALNERSSRSHSVLTVHVRGRELVSGSILRG-- 372
Query: 248 ASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLS 292
L+ VDLAGSER ++ G RLKE HIN+SL L VI L+
Sbjct: 373 -CLHLVDLAGSERVEKSEAVGERLKEAQHINRSLSALGDVISALA 504
>AV407795
Length = 424
Score = 73.9 bits (180), Expect = 6e-14
Identities = 52/140 (37%), Positives = 75/140 (53%), Gaps = 2/140 (1%)
Frame = +2
Query: 156 EIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGET 215
EIY + DLLN + L I +D ++ + L E D + ++ LI A R G T
Sbjct: 8 EIYGGKLFDLLN-DRKKLFIREDGKQQVCIVGLQEYCVSDVESIKELIERGNATRSTGTT 184
Query: 216 TLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQT--NTCGTRLKE 273
N++SSRSH I++L ++ + + L+F+DLAGSER + T N TR+ E
Sbjct: 185 GANEESSRSHAILQLAIKRSVDGNESKPPRPVGKLSFIDLAGSERGADTTDNDKQTRI-E 361
Query: 274 GSHINKSLLQLALVIRQLSS 293
G+ INKSLL L IR L +
Sbjct: 362 GAEINKSLLALKECIRALDN 421
>CB829109
Length = 514
Score = 67.4 bits (163), Expect = 6e-12
Identities = 45/127 (35%), Positives = 67/127 (52%), Gaps = 2/127 (1%)
Frame = +2
Query: 109 ATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNT--PDRDFVLKISALEIYNETVIDLL 166
AT FAYGQT SGKT+TM+ + A D+ + ++ F L +S EIY + DLL
Sbjct: 50 ATCFAYGQTGSGKTYTMQPLPLRAAEDLVRQLHRPVYQNQKFKLWLSYFEIYGGKLFDLL 229
Query: 167 NRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQ 226
+ + L + +D + + L E D Q ++ I A R G T N++SSRSH
Sbjct: 230 S-DRKKLCMREDGRQQVCIVGLQEFEVFDVQIVKEFIEKGNAARSTGSTGANEESSRSHA 406
Query: 227 IIRLTVE 233
I++L V+
Sbjct: 407 ILQLVVK 427
>TC15461 similar to UP|O24147 (O24147) Kinesin-like protein, partial (9%)
Length = 527
Score = 61.2 bits (147), Expect = 4e-10
Identities = 36/111 (32%), Positives = 60/111 (53%)
Frame = +3
Query: 287 VIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNA 346
VI LSSG HI YR KLT ++ SLGGNA+T + VSP S +++T N+L +A+
Sbjct: 9 VISALSSGGQ-HIPYRNHKLTMLMSDSLGGNAKTLMFVNVSPIESSLDETHNSLMYASRV 185
Query: 347 KEVINTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQR 397
+ ++N N+ E+ + + A + + E+D+E+++ R
Sbjct: 186 RSIVNDPSKNVSSKEIMRLKKQVAYWKEQAGRR-----GEEEDLEEIQEVR 323
>TC12682
Length = 583
Score = 52.0 bits (123), Expect(2) = 4e-08
Identities = 24/58 (41%), Positives = 39/58 (66%), Gaps = 4/58 (6%)
Frame = -1
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTSYTFDRVF 82
EE+I V+VR+RPLN KE A D+ W+C++D TI++++ + T+Y+FD+ F
Sbjct: 217 EERIHVSVRLRPLNDKELARNDVSEWECINDNTILYRSDFSASERSLYPTAYSFDKSF 44
Score = 22.3 bits (46), Expect(2) = 4e-08
Identities = 9/17 (52%), Positives = 12/17 (69%)
Frame = -2
Query: 80 RVFPPACSTQKVYDEGA 96
RV CST++VY+E A
Sbjct: 51 RVLGSDCSTRQVYEEAA 1
>TC15187 similar to UP|O24147 (O24147) Kinesin-like protein, partial (8%)
Length = 611
Score = 54.3 bits (129), Expect = 5e-08
Identities = 25/64 (39%), Positives = 39/64 (60%)
Frame = +2
Query: 299 ISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV 358
I YR KLT ++ SLGGNA+T + VSP S +++T N+L +A+ + +IN N+
Sbjct: 2 IPYRNHKLTMLMSDSLGGNAQTLMFVNVSPVESSLDETHNSLMYASRVRSIINDPSQNVA 181
Query: 359 EGEL 362
E+
Sbjct: 182 SKEI 193
>BP063922
Length = 537
Score = 43.9 bits (102), Expect = 7e-05
Identities = 33/94 (35%), Positives = 49/94 (52%)
Frame = +2
Query: 220 KSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINK 279
K + SH I+ T+ F+ SY + L+ VDLAGSE + + G R+ + H+ K
Sbjct: 266 KINVSHLIV--TIHIFYNNLITGENSY-SKLSLVDLAGSEGSITEDDSGERVTDLLHVMK 436
Query: 280 SLLQLALVIRQLSSGKSGHISYRTSKLTRILQSS 313
SL L V+ L+S K + Y S LT++L S
Sbjct: 437 SLSALGDVLSSLTS-KKDIVPYENSALTKVLADS 535
>AV421105
Length = 415
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/53 (37%), Positives = 34/53 (63%)
Frame = +2
Query: 299 ISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVIN 351
+ YR SKLTRILQ SLGG +R ++ ++P +++ +T+S A ++ + N
Sbjct: 5 VPYRESKLTRILQDSLGGTSRALMVACLNP--GEYQESVHTVSLAARSRHITN 157
>AV773555
Length = 460
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/76 (36%), Positives = 44/76 (57%), Gaps = 1/76 (1%)
Frame = -1
Query: 280 SLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNT 339
SL L+ VI L+ + HI +R SKLT +LQ +LGG+++T + +SP + ++ +
Sbjct: 460 SLSSLSDVIFALAQ-QDDHIPFRHSKLTYLLQPALGGDSKTVMFVNISPDPASAGESLCS 284
Query: 340 LSFATNAKE-VINTAR 354
L FA+ VI T R
Sbjct: 283 LRFASRVNACVIGTPR 236
>TC12031 homologue to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (7%)
Length = 740
Score = 37.7 bits (86), Expect = 0.005
Identities = 21/50 (42%), Positives = 32/50 (64%), Gaps = 2/50 (4%)
Frame = +2
Query: 302 RTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFAT--NAKEV 349
R SKLT +LQ LGG+++T + +SP S V ++ +L FA+ NA E+
Sbjct: 2 RNSKLTYLLQPCLGGDSKTLMFVNISPDQSSVGESLCSLRFASRVNACEI 151
>TC11319
Length = 1030
Score = 30.8 bits (68), Expect = 0.60
Identities = 23/95 (24%), Positives = 42/95 (44%), Gaps = 3/95 (3%)
Frame = +3
Query: 341 SFATNAKEVINTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLA 400
S++ + E I+ A+ N ++ HAG SLL +LK + + +L +R++A
Sbjct: 360 SWSWSLNEFISIAKENKENVDVPGSALNHAGESSLLVRSDLKDEIACERERNLHLEREVA 539
Query: 401 QCQLDLERRANKVQKGSSDYGP---SSQVVRCLSF 432
++ ++ SD P + QV C F
Sbjct: 540 SNEVPFDKTGLNASSDGSDTDPA*ENGQV*EC*CF 644
>AV775627
Length = 448
Score = 28.5 bits (62), Expect = 3.0
Identities = 17/48 (35%), Positives = 28/48 (57%), Gaps = 2/48 (4%)
Frame = -3
Query: 309 ILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFAT--NAKEVINTAR 354
I Q LGG+++T + +SP S V ++ +L FA+ NA E+ + R
Sbjct: 380 IEQPCLGGDSKTLMFVNISPDPSSVSESLCSLRFASRVNACEIGSARR 237
>TC18478 homologue to UP|UAP1_ARATH (O64765) Probable
UDP-N-acetylglucosamine pyrophosphorylase , partial
(19%)
Length = 559
Score = 28.5 bits (62), Expect = 3.0
Identities = 14/46 (30%), Positives = 22/46 (47%)
Frame = -2
Query: 83 PPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGI 128
PPA TQ+V+ ALS +S ++ F G SS + + +
Sbjct: 159 PPAAITQRVWSRRTSSFALSGVS*LDPFAFFTGANSSSRNISKSAV 22
>TC17598 weakly similar to GB|AAP68293.1|31711874|BT008854 At1g80360
{Arabidopsis thaliana;}, partial (27%)
Length = 562
Score = 27.7 bits (60), Expect = 5.0
Identities = 29/99 (29%), Positives = 48/99 (48%), Gaps = 7/99 (7%)
Frame = -1
Query: 252 FVDLAGSERASQTNTCG-TRLKEGSHINKSLLQLALVIRQLSSGKSG------HISYRTS 304
F+ +A ++ NTC TRLK +N + +Q+ RQL S +G ++S+ +
Sbjct: 529 FLSIALNDGIQNENTCKLTRLK----VNLTFIQM----RQLQSCSTGKWSFFLYVSFYSL 374
Query: 305 KLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFA 343
+ +SSLG P+L+H++ T TLS A
Sbjct: 373 DMR**TRSSLG------------PALTHLQPTLPTLSSA 293
>BI420256
Length = 524
Score = 27.3 bits (59), Expect = 6.6
Identities = 15/48 (31%), Positives = 28/48 (58%)
Frame = +3
Query: 366 EPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKV 413
+PE G+RS+L E E ++ ++K E++ +++ Q DL R K+
Sbjct: 318 DPEIDGIRSVLRESEKVLESLQKQEENM--LQEVTQKAKDLHDREYKL 455
>AU251440
Length = 331
Score = 26.9 bits (58), Expect = 8.6
Identities = 15/59 (25%), Positives = 25/59 (41%)
Frame = +2
Query: 430 LSFAEENELAIGKHTPERTETVSRQAMLKNLLASPDPSILVDEIQKLEHRQLQLCEDAN 488
+ + E + G + P E S + +K L+D + LEHR QLC + +
Sbjct: 98 MKYLERKQRCGGINVPAAAEKQSSDSEIKEAYFK---GTLLDRVASLEHRLFQLCVEVD 265
>BP041458
Length = 469
Score = 26.9 bits (58), Expect = 8.6
Identities = 24/83 (28%), Positives = 37/83 (43%), Gaps = 5/83 (6%)
Frame = -2
Query: 117 TSSGKTFTMRGITENAIRDIYECIKNTPDRDFVLKISAL-EIYNETVIDLLNRESGPLRI 175
T K T +N + Y N R+ +L++ + E E +I L RE PL+
Sbjct: 465 TPLDKFSTRSSALKNHLVQEYIDFLNNASREELLELKGIGEKMAEYIIGL--REESPLKS 292
Query: 176 LDDTEKVTV----VENLFEEVAR 194
L D EK+ + NLF + A+
Sbjct: 291 LSDLEKIGLSSKQAHNLFTKAAK 223
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.130 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,550,142
Number of Sequences: 28460
Number of extensions: 89200
Number of successful extensions: 402
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 395
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 397
length of query: 552
length of database: 4,897,600
effective HSP length: 95
effective length of query: 457
effective length of database: 2,193,900
effective search space: 1002612300
effective search space used: 1002612300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC124967.12


