

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124951.5 + phase: 0
(285 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
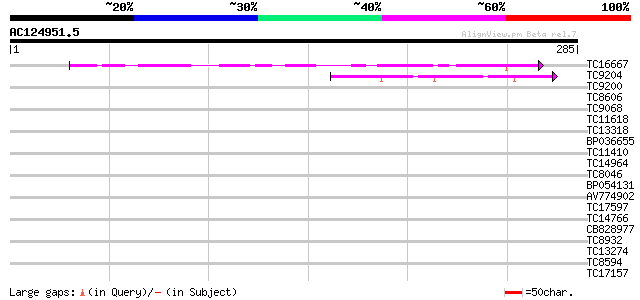
Score E
Sequences producing significant alignments: (bits) Value
TC16667 similar to UP|O64594 (O64594) F17O7.4 (AT1G70420/F17O7_4... 70 4e-13
TC9204 similar to PIR|T00986|T00986 yeast pheromone receptor-lik... 46 8e-06
TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, ... 36 0.009
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 32 0.095
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 32 0.12
TC11618 homologue to AAQ62405 (AAQ62405) At5g09680, partial (10%) 30 0.36
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 30 0.61
BP036655 29 0.80
TC11410 28 1.4
TC14964 weakly similar to UP|SOX_ARATH (Q9SJA7) Potential sarcos... 28 1.8
TC8046 similar to UP|Q7XJC2 (Q7XJC2) Adenine phosphoribosyltrans... 28 1.8
BP054131 28 2.3
AV774902 28 2.3
TC17597 similar to UP|AZF1_YEAST (P41696) Asparagine-rich zinc f... 28 2.3
TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associ... 28 2.3
CB828977 27 3.0
TC8932 similar to UP|Q7YY80 (Q7YY80) Serine repeat antigen, prob... 27 4.0
TC13274 weakly similar to GB|AAO24530.1|27808500|BT003098 At3g14... 27 5.2
TC8594 similar to UP|GCSH_PEA (P16048) Glycine cleavage system H... 27 5.2
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 27 5.2
>TC16667 similar to UP|O64594 (O64594) F17O7.4 (AT1G70420/F17O7_4), partial
(23%)
Length = 953
Score = 70.1 bits (170), Expect = 4e-13
Identities = 76/244 (31%), Positives = 108/244 (44%), Gaps = 6/244 (2%)
Frame = +2
Query: 31 NDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADVSPIFNRDERRNSDAMDI-SISLKN 89
N+V + E D+ + ++F FV D SPI D N I + +N
Sbjct: 194 NEVEDEEQVEEDDD--EEEEFSFVLVNP------DGSPISADDAFDNGQIRQIYPVFDQN 349
Query: 90 LLIGDEKPKLNGGGRRNSDAAEILYS-LKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFW 148
LL +SD + YS LKK+F+ +K +SS + P E+YC W
Sbjct: 350 LL--------------SSDGGDSHYSPLKKVFV--QKTGDSSSA------AAPEETYCEW 463
Query: 149 TPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVT 208
+PK++ KSNSTGSS WKF + RS SDGK++ +
Sbjct: 464 SPKTAV-----------------KSNSTGSS-----KVWKFRDVKLRSNSDGKDAFVFLN 577
Query: 209 PAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFY----GRKKETRVKSYLPYKK 264
E K + G VV KK+ A + KT + SA E Y +K+ + KSYLPY++
Sbjct: 578 STAAE--KASPTG---VVVKKVKAVKGKTTTSTSAHEKHYVLNRAKKESDKRKSYLPYRQ 742
Query: 265 ELIG 268
EL G
Sbjct: 743 ELFG 754
>TC9204 similar to PIR|T00986|T00986 yeast pheromone receptor-like protein
AR781 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (25%)
Length = 644
Score = 45.8 bits (107), Expect = 8e-06
Identities = 39/129 (30%), Positives = 66/129 (50%), Gaps = 15/129 (11%)
Frame = +2
Query: 162 SPMNSTIKCKKSNSTGSSSNTSSS--RWKFLS-LLRRSKSDGKESLNMVTPAKK------ 212
S +NS S+S+ SS++ SS +W+ LL RS S+G+ S P +K
Sbjct: 131 SDLNSAPATAASSSSLSSTSKGSSGRKWRLRDFLLFRSASEGRGS--STDPLRKFHLFYK 304
Query: 213 --ENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRKK----ETRVKSYLPYKKEL 266
E +K +S + + G + P +K P VSA E+ Y RK+ + + +++LPYK+ +
Sbjct: 305 KGEEVKASSSTSTSFRGSETPRLRRKEP--VSAHELHYARKRAETEDLKKRTFLPYKQGI 478
Query: 267 IGFSVGFNA 275
+G G +
Sbjct: 479 LGRLAGLGS 505
>TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(31%)
Length = 727
Score = 35.8 bits (81), Expect = 0.009
Identities = 21/47 (44%), Positives = 26/47 (54%)
Frame = -3
Query: 139 SIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
S P S FW SSS AS TS +S+ S S+ SSS++SSS
Sbjct: 374 SFPMGSRSFWIGSSSSSSASASTSSSSSSSSSSSSPSSSSSSSSSSS 234
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 32.3 bits (72), Expect = 0.095
Identities = 26/88 (29%), Positives = 39/88 (43%)
Frame = +1
Query: 100 NGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASP 159
N RR D+A ++Y LF+G +SS SSS S
Sbjct: 202 NNTNRRRLDSANVVYFF--LFLGGCSASSSSSS-------------------SSSSSLSS 318
Query: 160 KTSPMNSTIKCKKSNSTGSSSNTSSSRW 187
++P +S+ S+S+ SSS++SSS W
Sbjct: 319 SSTPSSSSSSPPSSSSSSSSSSSSSSSW 402
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 32.0 bits (71), Expect = 0.12
Identities = 23/59 (38%), Positives = 31/59 (51%)
Frame = -1
Query: 142 AESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDG 200
+ES CF +P SSS +S S+S+ SSS++SSS L L +R K DG
Sbjct: 180 SESGCFLSPSSSSSSSS--------------SSSSSSSSSSSSSSSSGLGLNKRLKPDG 46
>TC11618 homologue to AAQ62405 (AAQ62405) At5g09680, partial (10%)
Length = 514
Score = 30.4 bits (67), Expect = 0.36
Identities = 40/168 (23%), Positives = 70/168 (40%), Gaps = 9/168 (5%)
Frame = +2
Query: 106 NSDAAEILYSLKKLFIGNEKEK-WTSSEVE--DDLDSIPAESYCFWTPKSSSLIASPKTS 162
NS A+ +L L N + W SE++ DD S+ ++ + S
Sbjct: 20 NSIASSLLRPNYHLIAANSYQNLWL*SEMDTGDDFTFCQVSSHVDGDGSETNKLVSNIAD 199
Query: 163 PMNSTIKCKKSNSTGSSSNTSSS--RWK----FLSLLRRSKSDGKESLNMVTPAKKENLK 216
S+S+ SSSNTS++ WK S R+ + G S ++V A + +
Sbjct: 200 ISIKEESSNTSSSSSSSSNTSNTGLLWKDGLPNDSNSRKESTIGSLSFSVVNTASNQPSE 379
Query: 217 LNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRKKETRVKSYLPYKK 264
L+S K +P+ + +P S ++ +K R K +P++K
Sbjct: 380 LSS--------KLVPSNAENSPPQESPEQMISAKKPIVRTK--VPFEK 493
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 29.6 bits (65), Expect = 0.61
Identities = 17/41 (41%), Positives = 26/41 (62%)
Frame = -3
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSL 192
SSS +S +SP +S+ S+S+ SSS++SSS LS+
Sbjct: 364 SSSSSSSSSSSPSSSSSSSSSSSSSSSSSSSSSSSSSSLSV 242
Score = 27.3 bits (59), Expect = 3.0
Identities = 17/47 (36%), Positives = 26/47 (55%)
Frame = -3
Query: 139 SIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
S P+ S SSS +S +S +S+ S+S+ SSS++SSS
Sbjct: 406 SFPSTSTSSLPSSSSSSSSSSSSSSPSSSSSSSSSSSSSSSSSSSSS 266
>BP036655
Length = 556
Score = 29.3 bits (64), Expect = 0.80
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Frame = +1
Query: 147 FWTPKSSSLIASPKTSPMNS--TIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESL 204
F+TP SSS ++P +S +S ++ S S+ +SS+ SSS SL + S +
Sbjct: 193 FFTPISSSTFSTPHSSSSSSSFSVSISVSLSSSTSSSLSSSLISSSSLHSSTTSSSSSTT 372
Query: 205 NMVTPAKK 212
+ +P+ +
Sbjct: 373 SSSSPSSE 396
>TC11410
Length = 535
Score = 28.5 bits (62), Expect = 1.4
Identities = 17/59 (28%), Positives = 26/59 (43%)
Frame = +2
Query: 138 DSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRS 196
+++ E YC W + S + P P N + C S GS S++ + FL R S
Sbjct: 80 ETLNKERYCSWRERDISGVMHPSFDPQNQSC-CMPSLMAGSRLV*SNNLFMFLKCSRAS 253
>TC14964 weakly similar to UP|SOX_ARATH (Q9SJA7) Potential sarcosine oxidase
, partial (33%)
Length = 612
Score = 28.1 bits (61), Expect = 1.8
Identities = 16/34 (47%), Positives = 23/34 (67%)
Frame = +1
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
SS+ +SP++SP NST S +TG SS TS++
Sbjct: 373 SSTAASSPRSSPENST-----SRTTGLSSPTSTA 459
Score = 26.9 bits (58), Expect = 4.0
Identities = 19/39 (48%), Positives = 23/39 (58%)
Frame = +1
Query: 153 SSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS 191
SS IAS TSPMNS+ ++S SS S+SR LS
Sbjct: 334 SSRIASATTSPMNSS---TAASSPRSSPENSTSRTTGLS 441
>TC8046 similar to UP|Q7XJC2 (Q7XJC2) Adenine phosphoribosyltransferase
(Fragment), partial (85%)
Length = 1298
Score = 28.1 bits (61), Expect = 1.8
Identities = 17/43 (39%), Positives = 25/43 (57%), Gaps = 2/43 (4%)
Frame = +1
Query: 168 IKCKKSNSTGSSSNTSSSRWKFLSL--LRRSKSDGKESLNMVT 208
+ K +NST SSS++SSS +F + LR + S S NM +
Sbjct: 160 LSAKIANSTSSSSSSSSSSIRFQNSPPLRSTASSSSSSANMAS 288
>BP054131
Length = 500
Score = 27.7 bits (60), Expect = 2.3
Identities = 20/54 (37%), Positives = 26/54 (48%)
Frame = +3
Query: 156 IASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTP 209
I+S +S +S+ KS+S SSS+TSSS L LL K N P
Sbjct: 165 ISSSSSSIASSSSVYTKSSSISSSSSTSSSSSSILLLLLAFSWSFKPVFNTTVP 326
>AV774902
Length = 594
Score = 27.7 bits (60), Expect = 2.3
Identities = 17/40 (42%), Positives = 24/40 (59%), Gaps = 1/40 (2%)
Frame = -3
Query: 158 SPKTSPM-NSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRS 196
+PKTSP T++C SS++ S SRW ++LLR S
Sbjct: 259 TPKTSPYATETLQCH------SSAHDSRSRWPQINLLRCS 158
>TC17597 similar to UP|AZF1_YEAST (P41696) Asparagine-rich zinc finger
protein AZF1, partial (3%)
Length = 399
Score = 27.7 bits (60), Expect = 2.3
Identities = 16/46 (34%), Positives = 27/46 (57%)
Frame = +1
Query: 2 QNQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQ 47
QNQ QN+ +SS+ + P + ++NN N+ +++ N NS SQ
Sbjct: 184 QNQNQNQSSSSSPMNSPSPRSNASNNNNYNN-NNCMSSSNSNSISQ 318
>TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associated
complex alpha chain, partial (58%)
Length = 600
Score = 27.7 bits (60), Expect = 2.3
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = -1
Query: 146 CFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKF 189
CF P +S + P +S+ S+S+ SSS T SS W F
Sbjct: 234 CFDLPDASP--SCPSMLSSSSSSSSSSSSSSSSSSTTGSSSWIF 109
>CB828977
Length = 528
Score = 27.3 bits (59), Expect = 3.0
Identities = 22/76 (28%), Positives = 34/76 (43%), Gaps = 8/76 (10%)
Frame = +1
Query: 121 IGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPK-----TSPMNSTIKCKKSN- 174
+GN+K+KWT+ E D++ + K +++ P+ TS N +K K N
Sbjct: 52 MGNQKQKWTAEEE----DALHRGVQKYGAGKWKNILKDPEFAPSLTSRSNIDLKDKWRNL 219
Query: 175 --STGSSSNTSSSRWK 188
TG SN S K
Sbjct: 220 NVGTGQGSNVKSRTLK 267
>TC8932 similar to UP|Q7YY80 (Q7YY80) Serine repeat antigen, probable,
partial (13%)
Length = 914
Score = 26.9 bits (58), Expect = 4.0
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Frame = +3
Query: 162 SPMNST--IKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPA 210
S M ST I C + +S+ SSS++SSS K + + S S+ L + PA
Sbjct: 492 SAMRSTFQINCSQRSSSTSSSSSSSSLTKSIVISLPSSSENISLLARLGPA 644
>TC13274 weakly similar to GB|AAO24530.1|27808500|BT003098 At3g14430
{Arabidopsis thaliana;}, partial (78%)
Length = 450
Score = 26.6 bits (57), Expect = 5.2
Identities = 15/20 (75%), Positives = 15/20 (75%), Gaps = 1/20 (5%)
Frame = +2
Query: 152 SSSLIASPKTSPMN-STIKC 170
SSSLI S KTSP STIKC
Sbjct: 98 SSSLIFSSKTSPQGLSTIKC 157
>TC8594 similar to UP|GCSH_PEA (P16048) Glycine cleavage system H protein,
mitochondrial precursor , complete
Length = 742
Score = 26.6 bits (57), Expect = 5.2
Identities = 13/36 (36%), Positives = 22/36 (61%)
Frame = -1
Query: 154 SLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKF 189
+L +PK SP+ + + +GSS+NT+S RW +
Sbjct: 328 TLSTAPKPSPLVTGLP-----DSGSSTNTTSPRWSW 236
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 26.6 bits (57), Expect = 5.2
Identities = 16/46 (34%), Positives = 26/46 (55%)
Frame = -3
Query: 140 IPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
+P F +P SSS +S +S +S+ S+S+ SSS++S S
Sbjct: 903 LPLLLLLFPSPSSSSSSSSSSSSSSSSSSSSSSSSSSPSSSSSSIS 766
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.309 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,500,362
Number of Sequences: 28460
Number of extensions: 60219
Number of successful extensions: 579
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 451
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 510
length of query: 285
length of database: 4,897,600
effective HSP length: 89
effective length of query: 196
effective length of database: 2,364,660
effective search space: 463473360
effective search space used: 463473360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC124951.5


