

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
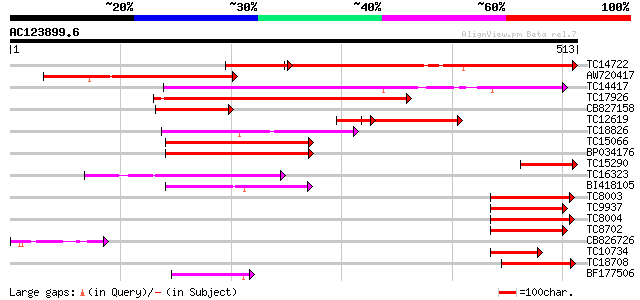
Query= AC123899.6 + phase: 0 /pseudo
(513 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14722 homologue to UP|Q9ZS12 (Q9ZS12) RNA helicase, partial (64%) 330 e-111
AW720417 295 2e-80
TC14417 homologue to UP|AAR23806 (AAR23806) Initiation factor eI... 185 1e-47
TC17926 similar to UP|IF43_NICPL (P41380) Eukaryotic initiation ... 173 7e-44
CB827158 137 3e-33
TC12619 homologue to UP|Q9ZS12 (Q9ZS12) RNA helicase, partial (23%) 117 4e-27
TC18826 similar to UP|Q9LUW5 (Q9LUW5) DEAD-box RNA helicase-like... 114 5e-26
TC15066 homologue to UP|Q8H1A5 (Q8H1A5) DEAD box RNA helicase, p... 110 4e-25
BP034176 107 6e-24
TC15290 UP|Q9ZS12 (Q9ZS12) RNA helicase, partial (10%) 106 1e-23
TC16323 similar to UP|Q9ZS06 (Q9ZS06) RNA helicase (Fragment), p... 91 5e-19
BI418105 85 2e-17
TC8003 homologue to PIR|S52023|S52023 translation initiation fac... 76 1e-14
TC9937 homologue to UP|IF43_NICPL (P41380) Eukaryotic initiation... 75 2e-14
TC8004 homologue to PIR|S52023|S52023 translation initiation fac... 74 4e-14
TC8702 homologue to UP|IF42_ARATH (P41377) Eukaryotic initiation... 73 1e-13
CB826726 67 9e-12
TC10734 homologue to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90 (AT5g... 65 2e-11
TC18708 similar to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90 (AT5g11... 64 5e-11
BF177506 60 1e-09
>TC14722 homologue to UP|Q9ZS12 (Q9ZS12) RNA helicase, partial (64%)
Length = 1448
Score = 330 bits (846), Expect(2) = e-111
Identities = 187/271 (69%), Positives = 203/271 (74%), Gaps = 6/271 (2%)
Frame = +3
Query: 249 KDDIMRLYQPVHLLVGTPGRILDLAKKGVCVL-KDCSMLVMDEADKLLSPEFQPSIEQLI 307
K DIMRLYQPVHLLVG + + ++ V K CSMLVMDEADKLLSPEFQPSIEQLI
Sbjct: 195 KYDIMRLYQPVHLLVGKARKEY*ILQRKVFAF*KHCSMLVMDEADKLLSPEFQPSIEQLI 374
Query: 308 QFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHC 367
QFL P RQILM+SAT+PVTVKDFK+RYLRKPY+INLMDELTLKGITQFYAFVEERQKVHC
Sbjct: 375 QFLSPNRQILMYSATYPVTVKDFKERYLRKPYVINLMDELTLKGITQFYAFVEERQKVHC 554
Query: 368 LNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP**SFPR 422
LNTLFSK I+ + F S E ++ YSC K + F
Sbjct: 555 LNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMLQDHRNRVFHD 719
Query: 423 LPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLIT 482
+ LFTRGIDIQAVNVVINFDFPKN+ETYLHRVGRSGR+GHLGLAVNLIT
Sbjct: 720 FRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKNAETYLHRVGRSGRYGHLGLAVNLIT 899
Query: 483 YEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
YEDRFNLYRIEQELGTEIKQIPP IDQAIYC
Sbjct: 900 YEDRFNLYRIEQELGTEIKQIPPHIDQAIYC 992
Score = 89.0 bits (219), Expect(2) = e-111
Identities = 47/61 (77%), Positives = 51/61 (83%)
Frame = +1
Query: 196 IPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRL 255
IPA EKID DNN+IQVVIL TRELA QTSQVC ELGKHL+IQVMVTTGGTSL+ I +
Sbjct: 34 IPA*EKIDHDNNVIQVVILDETRELASQTSQVCIELGKHLKIQVMVTTGGTSLESMISCV 213
Query: 256 Y 256
Y
Sbjct: 214 Y 216
>AW720417
Length = 554
Score = 295 bits (754), Expect = 2e-80
Identities = 151/185 (81%), Positives = 155/185 (83%), Gaps = 9/185 (4%)
Frame = +3
Query: 31 SNTGFQQRPHHQYQQQQQYVQRHMMQNQHQQHYQNQQQNQ---------QQNQQQQQQQQ 81
SN GFQ RPH Q QQQ QYVQRHMM NQ Q H Q QQNQ Q QQQQQQQQ
Sbjct: 3 SNPGFQPRPHQQQQQQHQYVQRHMMPNQQQYHQQQHQQNQPQYHQQHQHNQQQQQQQQQQ 182
Query: 82 WLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNE 141
WLRR QLGG DTNVVEEVEKTVQSE+ D SSQDWKARLK+PP DTRY+TEDVTATKGNE
Sbjct: 183 WLRRTQLGGN-DTNVVEEVEKTVQSESADPSSQDWKARLKIPPPDTRYQTEDVTATKGNE 359
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
FEDYFLKRELLMGIYEKGFE+PSPIQEESIPIALTGSDILARAKNGTGKTAAF IPALEK
Sbjct: 360 FEDYFLKRELLMGIYEKGFEKPSPIQEESIPIALTGSDILARAKNGTGKTAAFCIPALEK 539
Query: 202 IDQDN 206
ID DN
Sbjct: 540 IDHDN 554
>TC14417 homologue to UP|AAR23806 (AAR23806) Initiation factor eIF4A-15,
complete
Length = 1563
Score = 185 bits (470), Expect = 1e-47
Identities = 123/386 (31%), Positives = 195/386 (49%), Gaps = 21/386 (5%)
Frame = +2
Query: 140 NEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPAL 199
+ F+ L+ LL GIY GFERPS IQ+ I G D++ +A++GTGKTA F L
Sbjct: 206 DSFDAMGLQENLLRGIYAYGFERPSAIQQRGIVPFCKGLDVIQQAQSGTGKTATFCSGIL 385
Query: 200 EKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPV 259
+++D Q ++L PTRELA Q +V + LG +L ++V GGTS+++D L V
Sbjct: 386 QQLDYGLVQCQALVLAPTRELAQQIEKVMRALGDYLGVKVHACVGGTSVREDQRILQAGV 565
Query: 260 HLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMF 319
H +VGTPGR+ D+ ++ M V+DEAD++LS F+ I + Q LP Q+ +F
Sbjct: 566 HTVVGTPGRVFDMLRRQSLRPDSIKMFVLDEADEMLSRGFKDQIYDIFQLLPGKIQVGVF 745
Query: 320 SATFPVTVKDFKDRYLR-----------------KPYIINL-MDELTLKGITQFYAFVEE 361
SAT P + +++ K + +N+ +E L+ + Y +
Sbjct: 746 SATMPPEALEITRKFMNKPVRILVKRDELTLEGIKQFYVNVEKEEWKLETLCDLYETLAI 925
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFP 421
Q V +NT + + L +S+ H + +D+
Sbjct: 926 TQSVIFVNT------RRKVDWLTDKMRSNDHTVSATHGDMD----QNTRDII-------- 1051
Query: 422 RLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAV 478
+ ++ SR+ L RGID+Q V++VIN+D P E YLHR+GRSGRFG G+A+
Sbjct: 1052-MREFRSGSSRVLITTDLLARGIDVQQVSLVINYDLPTQPENYLHRIGRSGRFGRKGVAI 1228
Query: 479 NLITYEDRFNLYRIEQELGTEIKQIP 504
N +T +D L I++ ++++P
Sbjct: 1229NFVTLDDSRMLSDIQKFYNVTVEELP 1306
>TC17926 similar to UP|IF43_NICPL (P41380) Eukaryotic initiation factor 4A-3
(eIF4A-3) (eIF-4A-3), partial (72%)
Length = 982
Score = 173 bits (438), Expect = 7e-44
Identities = 91/234 (38%), Positives = 144/234 (60%), Gaps = 1/234 (0%)
Frame = +2
Query: 131 TEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGK 190
TE V A FE+ +K +LL GIY+ GFE+PS IQ+ ++ + G D++A+A++GTGK
Sbjct: 146 TEGVKAI--GSFEEMGIKDDLLRGIYQYGFEKPSAIQQRAVTPIIQGRDVIAQAQSGTGK 319
Query: 191 TAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKD 250
T+ ++ + +D +QV+IL PTRELA QT +V +G + IQ GG S+ +
Sbjct: 320 TSMIALTVCQVVDTSVREVQVLILSPTRELASQTEKVILAVGDFINIQAHACVGGKSVGE 499
Query: 251 DIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFL 310
DI +L VH++ GTPGR+ D+ K+ + MLV+DE+D++LS F+ I + ++L
Sbjct: 500 DIRKLEHGVHVVSGTPGRVCDMIKRRTLRTRTIKMLVLDESDEMLSRGFKDQIYDVYRYL 679
Query: 311 PPTRQILMFSATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQ 363
PP Q+ + SAT P + + ++++ P I+ DELTL+GI QF+ VE +
Sbjct: 680 PPDLQVCLISATLPHEILEITNKFMTDPVRILVKRDELTLEGIKQFFVAVEREE 841
>CB827158
Length = 213
Score = 137 bits (346), Expect = 3e-33
Identities = 68/70 (97%), Positives = 69/70 (98%)
Frame = +2
Query: 133 DVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTA 192
DVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTA
Sbjct: 2 DVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTA 181
Query: 193 AFSIPALEKI 202
AF IPALEK+
Sbjct: 182 AFCIPALEKV 211
>TC12619 homologue to UP|Q9ZS12 (Q9ZS12) RNA helicase, partial (23%)
Length = 346
Score = 117 bits (293), Expect = 4e-27
Identities = 58/91 (63%), Positives = 66/91 (71%)
Frame = +3
Query: 319 FSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANK 378
F FPVTVKDFKDR+L KPY+INLMDELTLKGITQFYA + L AANK
Sbjct: 72 FQQHFPVTVKDFKDRFLHKPYVINLMDELTLKGITQFYAVCGRETESPLLKYFIF*AANK 251
Query: 379 PIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC 409
P++H LQF +S *TPC ENH* VFMFL++C
Sbjct: 252 PVNHFLQFSESG*TPCQENH*AWVFMFLHTC 344
Score = 55.1 bits (131), Expect = 3e-08
Identities = 27/36 (75%), Positives = 29/36 (80%)
Frame = +2
Query: 296 SPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFK 331
SPEFQPSI+QLIQFLP RQILMFSATF K F+
Sbjct: 2 SPEFQPSIQQLIQFLPRNRQILMFSATFSCYCKGFQ 109
>TC18826 similar to UP|Q9LUW5 (Q9LUW5) DEAD-box RNA helicase-like protein,
partial (29%)
Length = 782
Score = 114 bits (284), Expect = 5e-26
Identities = 65/184 (35%), Positives = 102/184 (55%), Gaps = 6/184 (3%)
Frame = +3
Query: 138 KGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIP 197
+G E + + E++ + +KG + PIQ + A+ G D++ RA+ GTGKT AF IP
Sbjct: 237 EGLEIKKLGISEEIVSALAKKGISKLFPIQRAVLEPAMQGRDMIGRARTGTGKTLAFGIP 416
Query: 198 ALEKIDQDN------NIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDD 251
++KI + N + L PTRELA Q + + +L + GGT +
Sbjct: 417 IMDKIIKFNAKHGRGRDPLALALAPTRELAKQVEKEFYDSAPNLD--TICVNGGTPISQQ 590
Query: 252 IMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLP 311
+ +L V ++VGTPGRI+DL +G LK+ +V+DEAD++L FQ +E+++ LP
Sbjct: 591 MRQLDYGVDVVVGTPGRIIDLLNRGALNLKEVQFMVLDEADQMLQIGFQEDVEKILDRLP 770
Query: 312 PTRQ 315
P RQ
Sbjct: 771 PKRQ 782
>TC15066 homologue to UP|Q8H1A5 (Q8H1A5) DEAD box RNA helicase, partial
(44%)
Length = 699
Score = 110 bits (276), Expect = 4e-25
Identities = 56/134 (41%), Positives = 84/134 (61%)
Frame = +3
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+ L+ LL GIY GFE+PS IQ+ I G D++ +A++GTGKTA F L++
Sbjct: 282 FDAMGLQENLLRGIYAYGFEKPSAIQQRGIVPFCKGLDVIQQAQSGTGKTATFCSGILQQ 461
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
+D Q ++L PTRELA Q +V + LG +L ++V GGTS+++D L VH+
Sbjct: 462 LDYSLTECQALVLAPTRELAQQIEKVMRALGDYLGVKVHACVGGTSVREDQRILSSGVHV 641
Query: 262 LVGTPGRILDLAKK 275
+VGTPGR+ D+ ++
Sbjct: 642 VVGTPGRVFDMLRR 683
>BP034176
Length = 604
Score = 107 bits (266), Expect = 6e-24
Identities = 55/134 (41%), Positives = 82/134 (61%)
Frame = +2
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+ L LL GIY GFERPS IQ+ G D++ +A++GTGKTA F L++
Sbjct: 188 FDSMGLHENLLRGIYAYGFERPSAIQQRGXVPFCKGLDVIQQAQSGTGKTATFCSGILQQ 367
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
++ + Q ++L PTRELA Q +V + LG +L ++V GGTS+++D L VH
Sbjct: 368 LNYELVQCQALVLAPTRELAQQIEKVMRALGDYLGVKVHACVGGTSVREDQRILQAGVHT 547
Query: 262 LVGTPGRILDLAKK 275
+VGTPGR+ D+ ++
Sbjct: 548 VVGTPGRVFDMLRR 589
>TC15290 UP|Q9ZS12 (Q9ZS12) RNA helicase, partial (10%)
Length = 966
Score = 106 bits (264), Expect = 1e-23
Identities = 50/51 (98%), Positives = 50/51 (98%)
Frame = +1
Query: 463 HRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
HRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPP IDQAIYC
Sbjct: 1 HRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPHIDQAIYC 153
>TC16323 similar to UP|Q9ZS06 (Q9ZS06) RNA helicase (Fragment), partial
(35%)
Length = 580
Score = 90.9 bits (224), Expect = 5e-19
Identities = 62/183 (33%), Positives = 91/183 (48%), Gaps = 1/183 (0%)
Frame = +3
Query: 68 QNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADT 127
Q Q+Q ++ L+R +G DT EE E D +D KA PA
Sbjct: 27 QKQKQI*GLVRRSSRLQRGSMGELKDTEAYEE-------ELIDYEEEDEKAPDSAKPAAE 185
Query: 128 RYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNG 187
+ + + F D+ LK ELL I + GFE PS +Q E IP A+ G D++ +AK+G
Sbjct: 186 TAK-KGYVGIHSSGFRDFLLKPELLRAIVDSGFEHPSEVQHECIPQAILGMDVICQAKSG 362
Query: 188 TGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHL-QIQVMVTTGGT 246
GKTA F + +L++ID + ++L TRELA Q + +L I+V V GG
Sbjct: 363 MGKTAVFVLSSLQQIDPVPGQVAALVLCHTRELAYQICHEFERFSTYLPDIKVAVFYGGV 542
Query: 247 SLK 249
++K
Sbjct: 543 NIK 551
>BI418105
Length = 600
Score = 85.1 bits (209), Expect = 2e-17
Identities = 51/139 (36%), Positives = 76/139 (53%), Gaps = 6/139 (4%)
Frame = +3
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
FED ++ I ++G+E+P+ IQ +++P+ L+G DI+ AK G+GKTAAF +P +
Sbjct: 162 FEDCGFPPPVMSAIKKQGYEKPTSIQCQALPVVLSGRDIIGIAKTGSGKTAAFVLPMIVH 341
Query: 202 IDQDNNIIQV------VILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRL 255
I D +Q VI PTRELA Q K+ K I+V GG S + L
Sbjct: 342 I-MDQPELQKEEGPIGVICAPTRELAQQIYLESKKFAKSYGIRVSAVYGGMSKLEQFKEL 518
Query: 256 YQPVHLLVGTPGRILDLAK 274
++V TPGR++D+ K
Sbjct: 519 KAGCEIVVATPGRLIDMLK 575
>TC8003 homologue to PIR|S52023|S52023 translation initiation factor
eIF-4A.14 - common tobacco {Nicotiana tabacum;} ,
partial (34%)
Length = 708
Score = 75.9 bits (185), Expect = 1e-14
Identities = 33/76 (43%), Positives = 51/76 (66%)
Frame = +1
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
L RGID+Q V++VIN+D P E YLHR+GRSGRFG G+A+N +T +D L+ I++
Sbjct: 190 LLARGIDVQQVSLVINYDLPTQPENYLHRIGRSGRFGRKGVAINFVTKDDERMLFDIQKF 369
Query: 496 LGTEIKQIPPFIDQAI 511
++++P I + +
Sbjct: 370 YNVVVEELPSNIAELL 417
>TC9937 homologue to UP|IF43_NICPL (P41380) Eukaryotic initiation factor
4A-3 (eIF4A-3) (eIF-4A-3), partial (28%)
Length = 651
Score = 75.5 bits (184), Expect = 2e-14
Identities = 33/69 (47%), Positives = 49/69 (70%)
Frame = +2
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
++ RG+D+Q V++VIN+D P N E Y+HR+GRSGRFG G+A+N + +D L IEQ
Sbjct: 119 VWARGLDVQQVSLVINYDLPNNRELYIHRIGRSGRFGRKGVAINFVKSDDIKILRDIEQY 298
Query: 496 LGTEIKQIP 504
T+I ++P
Sbjct: 299 YSTQIDEMP 325
>TC8004 homologue to PIR|S52023|S52023 translation initiation factor
eIF-4A.14 - common tobacco {Nicotiana tabacum;} ,
partial (20%)
Length = 607
Score = 74.3 bits (181), Expect = 4e-14
Identities = 32/76 (42%), Positives = 51/76 (67%)
Frame = +1
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
L RGID+Q V++VIN+D P E YLHR+GRSGRFG G+A+N ++ +D L+ I++
Sbjct: 25 LLARGIDVQQVSLVINYDLPTQPENYLHRIGRSGRFGRKGVAINFVSKDDERMLFDIQKF 204
Query: 496 LGTEIKQIPPFIDQAI 511
++++P I + +
Sbjct: 205 YNVVVEELPSNIAELL 252
>TC8702 homologue to UP|IF42_ARATH (P41377) Eukaryotic initiation factor
4A-2 (eIF4A-2) (eIF-4A-2), partial (32%)
Length = 595
Score = 73.2 bits (178), Expect = 1e-13
Identities = 32/69 (46%), Positives = 47/69 (67%)
Frame = +2
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
L RGID+Q V++VIN+D P E YLHR+GRSGRFG G+A+N +T +D L I++
Sbjct: 164 LLARGIDVQQVSLVINYDLPTQPENYLHRIGRSGRFGRKGVAINFVTLDDSRMLSDIQKF 343
Query: 496 LGTEIKQIP 504
++++P
Sbjct: 344 YHVTVEELP 370
>CB826726
Length = 505
Score = 66.6 bits (161), Expect = 9e-12
Identities = 48/101 (47%), Positives = 50/101 (48%), Gaps = 12/101 (11%)
Frame = +2
Query: 1 MNNNNRG-------RYP-----PGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQ 48
MNNNN RYP PGIG GRGG G N N GFQ R +Q QQQQQ
Sbjct: 236 MNNNNNNNNVNRANRYPLHPPPPGIGYGRGG----GPNPNPNPNQGFQPRLPYQQQQQQQ 403
Query: 49 YVQRHMMQNQHQQHYQNQQQNQQQNQQQQQQQQWLRRNQLG 89
QQHY Q+ QQQQQQQWLRR QLG
Sbjct: 404 -----------QQHYV-----QRNLAQQQQQQQWLRRAQLG 478
>TC10734 homologue to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90
(AT5g11170/F2I11_60), partial (15%)
Length = 513
Score = 65.5 bits (158), Expect = 2e-11
Identities = 28/47 (59%), Positives = 38/47 (80%)
Frame = +3
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLIT 482
L RGIDI+ VN+VIN+D P +++TYLHRVGR+GRFG GLA+ ++
Sbjct: 363 LVGRGIDIERVNIVINYDMPDSADTYLHRVGRAGRFGTKGLAITFVS 503
>TC18708 similar to UP|Q93VJ8 (Q93VJ8) AT5g11200/F2I11_90
(AT5g11170/F2I11_60), partial (16%)
Length = 582
Score = 64.3 bits (155), Expect = 5e-11
Identities = 30/68 (44%), Positives = 46/68 (67%), Gaps = 1/68 (1%)
Frame = -2
Query: 446 VNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYE-DRFNLYRIEQELGTEIKQIP 504
VN+VIN+D P +++TYLHR+GR+GRFG GLA+ ++ D L +++ +IK++P
Sbjct: 581 VNIVINYDMPDSADTYLHRLGRAGRFGTKGLAITFVSSSGDSEVLNQVQSRFEVDIKELP 402
Query: 505 PFIDQAIY 512
ID A Y
Sbjct: 401 EPIDTATY 378
>BF177506
Length = 447
Score = 59.7 bits (143), Expect = 1e-09
Identities = 33/83 (39%), Positives = 49/83 (58%), Gaps = 8/83 (9%)
Frame = +2
Query: 147 LKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDN 206
L +ELL + + G++ PSPIQ +IP+ L D++ A+ G+GKTAAF +P L I +
Sbjct: 197 LSQELLKSVEKAGYKTPSPIQMAAIPLGLQQRDVIGVAETGSGKTAAFVLPMLSYITRLP 376
Query: 207 NIIQ--------VVILVPTRELA 221
I + V++ PTRELA
Sbjct: 377 PISEENEAEGPYAVVMAPTRELA 445
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,020,659
Number of Sequences: 28460
Number of extensions: 107875
Number of successful extensions: 3085
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 1804
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2361
length of query: 513
length of database: 4,897,600
effective HSP length: 94
effective length of query: 419
effective length of database: 2,222,360
effective search space: 931168840
effective search space used: 931168840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC123899.6


