

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123596.12 - phase: 0
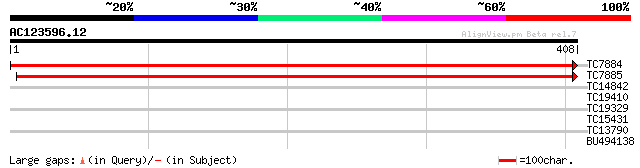
(408 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7884 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4... 734 0.0
TC7885 similar to UP|RL4B_ARATH (Q9SF40) 60S ribosomal protein L... 731 0.0
TC14842 similar to UP|RK4_TOBAC (O80361) 50S ribosomal protein L... 32 0.25
TC19410 weakly similar to AAR24668 (AAR24668) At1g24040, partial... 28 2.1
TC19329 similar to PIR|T52134|T52134 Zwille protein [imported] -... 28 2.8
TC15431 similar to GB|AAO63355.1|28950863|BT005291 At4g39540 {Ar... 28 3.6
TC13790 weakly similar to UP|Q8VW43 (Q8VW43) Proline dehydrogena... 27 6.2
BU494138 27 8.1
>TC7884 similar to UP|RL4_PRUAR (Q9XF97) 60S ribosomal protein L4 (L1),
partial (97%)
Length = 1476
Score = 734 bits (1896), Expect = 0.0
Identities = 365/408 (89%), Positives = 390/408 (95%)
Frame = +1
Query: 1 MAAAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 60
++ AAAAARPLVT+Q LE +M D+ T+PIPDVMRASIRPDIVNFVHSNISKNSRQPYA
Sbjct: 28 ISMAAAAARPLVTVQALEGDMVADAAATIPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 207
Query: 61 VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR 120
VSR+AGHQTSAESWGTGRAVSRIPRV GGGTHRAGQ AFGNMCRGGRMFAPTRIWR+WHR
Sbjct: 208 VSRRAGHQTSAESWGTGRAVSRIPRVPGGGTHRAGQGAFGNMCRGGRMFAPTRIWRRWHR 387
Query: 121 KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQ 180
KINVNQKR+A+VSAIAASA+PSLVLARGHRIETVPEFPLVV D+AEGVEK+KEAIKVLKQ
Sbjct: 388 KINVNQKRHALVSAIAASAVPSLVLARGHRIETVPEFPLVVSDSAEGVEKSKEAIKVLKQ 567
Query: 181 VGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNV 240
+GAF DAEKAKDS GIRPGKGKMRNRRYISR+GPLIVYGT+GAKAVKAFRNIPGVE+ NV
Sbjct: 568 IGAFPDAEKAKDSTGIRPGKGKMRNRRYISRRGPLIVYGTEGAKAVKAFRNIPGVEVANV 747
Query: 241 ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRI 300
ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYG+FDK SEKKKGY+LPR+KMVNSDL RI
Sbjct: 748 ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGSFDKPSEKKKGYLLPRSKMVNSDLARI 927
Query: 301 INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKE 360
INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEA+R+++KKE
Sbjct: 928 INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAQRLQAKKE 1107
Query: 361 KFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
K DK RKIVSKEEASAIKAAGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 1108KLDKMRKIVSKEEASAIKAAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 1251
>TC7885 similar to UP|RL4B_ARATH (Q9SF40) 60S ribosomal protein L4-2 (L1),
complete
Length = 1554
Score = 731 bits (1886), Expect = 0.0
Identities = 361/403 (89%), Positives = 387/403 (95%)
Frame = +3
Query: 6 AAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSRKA 65
AAARPLVT+Q LE +M D+ +T+PIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSR+A
Sbjct: 84 AAARPLVTVQALEGDMVADAASTIPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSRRA 263
Query: 66 GHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINVN 125
GHQTSAESWGTGRAVSRIPRV GGGTHRAGQ AFGNMCRGGRMFAPT+IWR+WHRKINVN
Sbjct: 264 GHQTSAESWGTGRAVSRIPRVPGGGTHRAGQGAFGNMCRGGRMFAPTKIWRRWHRKINVN 443
Query: 126 QKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAFA 185
QKR+A+VSAIAASA+PSLVLARGHRIETVPEFPLVV D+AEGVEK+KEAIKVLKQ+GAF
Sbjct: 444 QKRHALVSAIAASAVPSLVLARGHRIETVPEFPLVVSDSAEGVEKSKEAIKVLKQIGAFP 623
Query: 186 DAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNVERLNL 245
DAEKAKDS GIRPGKGKMRNRRYISR+GPL+VYGT+GAKAVKAFRNIPGVE+ NVERLNL
Sbjct: 624 DAEKAKDSTGIRPGKGKMRNRRYISRRGPLVVYGTEGAKAVKAFRNIPGVEVANVERLNL 803
Query: 246 LKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSDE 305
LKLAPGGHLGRFVIWTKSAFEKLDSIYG+FDK SEKKKGY+LPR+KMVNSDL RIINSDE
Sbjct: 804 LKLAPGGHLGRFVIWTKSAFEKLDSIYGSFDKPSEKKKGYLLPRSKMVNSDLARIINSDE 983
Query: 306 VQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDKK 365
VQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKR ALLAEA+R+++KKEK DKK
Sbjct: 984 VQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRAALLAEAQRLQAKKEKLDKK 1163
Query: 366 RKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
RKIVSKEEASAIKAAGKAWY+TMVSDSDY EFDNFSKWLGVSQ
Sbjct: 1164RKIVSKEEASAIKAAGKAWYHTMVSDSDYTEFDNFSKWLGVSQ 1292
>TC14842 similar to UP|RK4_TOBAC (O80361) 50S ribosomal protein L4,
chloroplast precursor (R-protein L4), partial (51%)
Length = 618
Score = 31.6 bits (70), Expect = 0.25
Identities = 26/81 (32%), Positives = 37/81 (45%), Gaps = 10/81 (12%)
Frame = +3
Query: 75 GTGRAVSRIPRVAGGGTHRAGQAAFGNMCRG----------GRMFAPTRIWRKWHRKINV 124
GT +R V GGG Q G RG G +F P R W KIN
Sbjct: 342 GTASTKTRA-EVRGGGKKPYPQKKTGRARRGSNRTPLRPGGGVIFGPRP--RDWSIKINR 512
Query: 125 NQKRYAVVSAIAASAIPSLVL 145
+KR A+ +A+A++A ++V+
Sbjct: 513 KEKRLAISTAVASAAANTIVV 575
>TC19410 weakly similar to AAR24668 (AAR24668) At1g24040, partial (28%)
Length = 606
Score = 28.5 bits (62), Expect = 2.1
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 3/39 (7%)
Frame = +2
Query: 65 AGHQTSAESWGTGRAVSRIPRVAGGGTHR---AGQAAFG 100
AG + W TG V R+ VAGG R +G+A FG
Sbjct: 395 AGARGGRRRWVTGGVVRRVDAVAGGLCQRVGVSGEAVFG 511
>TC19329 similar to PIR|T52134|T52134 Zwille protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(7%)
Length = 623
Score = 28.1 bits (61), Expect = 2.8
Identities = 15/46 (32%), Positives = 23/46 (49%)
Frame = -3
Query: 12 VTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQ 57
VT T+ S + + SP LP+P +RP VH+ + + RQ
Sbjct: 195 VTFGTMSSTLWSHSPAALPVPHRNELEMRPS----VHNMLEEQQRQ 70
>TC15431 similar to GB|AAO63355.1|28950863|BT005291 At4g39540 {Arabidopsis
thaliana;}, partial (39%)
Length = 757
Score = 27.7 bits (60), Expect = 3.6
Identities = 8/25 (32%), Positives = 16/25 (64%)
Frame = +2
Query: 98 AFGNMCRGGRMFAPTRIWRKWHRKI 122
+ GN+C G +F W++WH+++
Sbjct: 74 SIGNICTRGLVFGWMYQWKRWHKEL 148
>TC13790 weakly similar to UP|Q8VW43 (Q8VW43) Proline dehydrogenase, partial
(3%)
Length = 423
Score = 26.9 bits (58), Expect = 6.2
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = -1
Query: 88 GGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRK 121
G HR G+ +GN RGG AP +WR+ R+
Sbjct: 315 GKTRHRRGRRRWGNEGRGG---APR*LWRRSRRE 223
>BU494138
Length = 518
Score = 26.6 bits (57), Expect = 8.1
Identities = 14/47 (29%), Positives = 25/47 (52%)
Frame = -3
Query: 8 ARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKN 54
A PL+T T + + + NT+PIP + R + P + + + +KN
Sbjct: 381 ATPLLTFSTKPATIMDRTENTMPIPILWRRVMPPSCLVILLTAGTKN 241
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,445,541
Number of Sequences: 28460
Number of extensions: 61751
Number of successful extensions: 391
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 391
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 391
length of query: 408
length of database: 4,897,600
effective HSP length: 92
effective length of query: 316
effective length of database: 2,279,280
effective search space: 720252480
effective search space used: 720252480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123596.12


