

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.9 + phase: 0 /pseudo
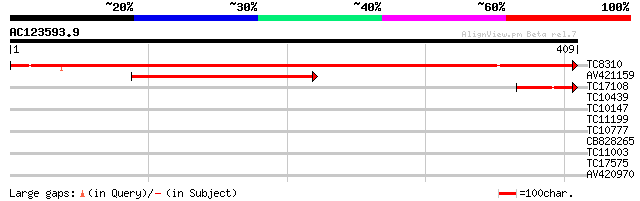
(409 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8310 UP|Q9SPM8 (Q9SPM8) Nod factor binding lectin-nucleotide p... 585 e-168
AV421159 179 1e-45
TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, pa... 53 8e-08
TC10439 similar to GB|AAO39907.1|28372850|BT003679 At1g67250 {Ar... 30 0.96
TC10147 weakly similar to GB|AAO39907.1|28372850|BT003679 At1g67... 28 2.1
TC11199 28 2.1
TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor S... 28 3.6
CB828265 27 4.7
TC11003 similar to UP|MYOP_MESCR (O49071) Inositol-1(or 4)-monop... 27 6.2
TC17575 27 6.2
AV420970 27 8.1
>TC8310 UP|Q9SPM8 (Q9SPM8) Nod factor binding lectin-nucleotide
phosphohydrolase, complete
Length = 1489
Score = 585 bits (1509), Expect = e-168
Identities = 288/411 (70%), Positives = 341/411 (82%), Gaps = 2/411 (0%)
Frame = +1
Query: 1 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIF--QKQETISSYAVVFDAGSTGSRI 58
M+FLI+L+T V +L MPAI+SSQYLGNN+L NRKI + QE ++SYAV+FDAGSTGSR+
Sbjct: 43 MDFLISLMTFVFML-MPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRV 219
Query: 59 HVYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHK 118
HVY+FDQNLDLL + ++EF++ + PGLSSYA +PE+AA+SLIPLL++AENVVP+
Sbjct: 220 HVYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPN 399
Query: 119 TPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVN 178
TP++LGATAGLRLL G+A+E ILQAVRDM SNRS NVQ DAVSI+DGTQEGSYLWVT+N
Sbjct: 400 TPVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTIN 579
Query: 179 YALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYD 238
Y LG LGK++TKTVGV+DLGGGSVQM YAVS+ TAKNAPKV +G DPYIKKLVL+GK YD
Sbjct: 580 YLLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYD 759
Query: 239 LYVHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCK 298
LYVHSYL +GREA RAEI KV S NPC+LAGFDG YTY+G E+K APASG+N N C+
Sbjct: 760 LYVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCR 939
Query: 299 KIIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKI 358
KI KALK+N PCPYQNCTFGGIWNGGGG+GQK+LF +SSF+YL EDVG+ K PN KI
Sbjct: 940 KIALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVNK-PNAKI 1116
Query: 359 RPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
RPVDL + AK AC N EDAKS YP L +K+ YVC+DL+Y Y LLVDGF
Sbjct: 1117RPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGF 1269
>AV421159
Length = 410
Score = 179 bits (453), Expect = 1e-45
Identities = 85/134 (63%), Positives = 109/134 (80%)
Frame = +2
Query: 89 YANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAGLRLLNGDASEKILQAVRDMF 148
YA +P+QAA+SL+ LL +AE+VVP +L KT +R+GATAGLR L GDAS++ILQAVRD+
Sbjct: 5 YAQNPQQAAESLVSLLDKAESVVPRELRSKTQVRVGATAGLRALEGDASDRILQAVRDLL 184
Query: 149 SNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMDLGGGSVQMAYAV 208
RS+ + DAV+++DG QEG++ WVT+NY LGNLGK Y+KTVGV+DLGGGSVQMAYA+
Sbjct: 185 KKRSSLKSEADAVTVLDGVQEGAFQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAI 364
Query: 209 SKKTAKNAPKVADG 222
S+ A APKV DG
Sbjct: 365 SETEAAQAPKVTDG 406
>TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (16%)
Length = 573
Score = 53.1 bits (126), Expect = 8e-08
Identities = 24/44 (54%), Positives = 29/44 (65%)
Frame = +1
Query: 366 EAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
+ K C DAKSTYP + N+ Y+CMDL+YQY LLVDGF
Sbjct: 1 QLSKLCQTKLGDAKSTYPHIEDGNLP-YICMDLVYQYTLLVDGF 129
>TC10439 similar to GB|AAO39907.1|28372850|BT003679 At1g67250 {Arabidopsis
thaliana;}, partial (59%)
Length = 461
Score = 29.6 bits (65), Expect = 0.96
Identities = 13/44 (29%), Positives = 22/44 (49%)
Frame = +1
Query: 85 GLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAG 128
G Y +DP +++E + P+D+HH +RLG + G
Sbjct: 322 GFEDYLSDP-----------RESETIRPLDMHHGMEVRLGLSKG 420
>TC10147 weakly similar to GB|AAO39907.1|28372850|BT003679 At1g67250
{Arabidopsis thaliana;}, partial (96%)
Length = 622
Score = 28.5 bits (62), Expect = 2.1
Identities = 14/44 (31%), Positives = 20/44 (44%)
Frame = +3
Query: 85 GLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAG 128
G NDP ++ SL P+D+HH +RLG + G
Sbjct: 372 GFEDCLNDPRESKTSL-----------PVDMHHGMEVRLGLSKG 470
>TC11199
Length = 743
Score = 28.5 bits (62), Expect = 2.1
Identities = 16/50 (32%), Positives = 28/50 (56%)
Frame = +3
Query: 221 DGVDPYIKKLVLKGKPYDLYVHSYLHFGREASRAEIMKVTRSSPNPCLLA 270
+ + P+I+K + GKP L+ H HF R + +EI +V ++ +LA
Sbjct: 147 EDLGPFIRKAFISGKPESLH-HHLRHFAR-SKESEIEEVCKAHYQDFILA 290
>TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor SF2-like
protein, partial (46%)
Length = 597
Score = 27.7 bits (60), Expect = 3.6
Identities = 16/50 (32%), Positives = 23/50 (46%)
Frame = +1
Query: 291 GANFNGCKKIIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFF 340
G NF+GC+ + A P +GG GGGG G + + +S F
Sbjct: 436 GYNFDGCRLRVELAHGGRGPTSSDRRGYGGGGGGGGGGGGRIGLSRNSEF 585
>CB828265
Length = 549
Score = 27.3 bits (59), Expect = 4.7
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 4/51 (7%)
Frame = +3
Query: 361 VDLVSEAKKACA-LNFEDAKSTYP---FLAKKNIASYVCMDLIYQYVLLVD 407
VDLV+E KAC L F K YP F+ ++ A++ +L + +VD
Sbjct: 195 VDLVTETDKACEDLIFNHLKQLYPAHKFIGEETSAAFGTTELTDEPTWIVD 347
>TC11003 similar to UP|MYOP_MESCR (O49071) Inositol-1(or 4)-monophosphatase
(IMPase) (IMP) (Inositol monophosphatase) , partial
(54%)
Length = 553
Score = 26.9 bits (58), Expect = 6.2
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Frame = +2
Query: 361 VDLVSEAKKAC-ALNFEDAKSTYP---FLAKKNIASYVCMDLIYQYVLLVD 407
VDLV+E KAC L F K YP F+ ++ A++ +L +VD
Sbjct: 200 VDLVTETDKACEELIFTQLKQQYPTHKFIGEETTAAFGTTELTDDPTWIVD 352
>TC17575
Length = 729
Score = 26.9 bits (58), Expect = 6.2
Identities = 9/24 (37%), Positives = 16/24 (66%)
Frame = +3
Query: 105 QQAENVVPIDLHHKTPIRLGATAG 128
++ E +P+D+HH +RLG + G
Sbjct: 519 REPETSLPLDMHHGMEVRLGLSKG 590
>AV420970
Length = 188
Score = 26.6 bits (57), Expect = 8.1
Identities = 16/45 (35%), Positives = 22/45 (48%)
Frame = +3
Query: 163 IIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMDLGGGSVQMAYA 207
+ DGT + L+ T A GN+G +V + DL GG V A
Sbjct: 3 LTDGTTLSTTLFQTTGTATGNVG-----SVNITDLKGGKVAFGSA 122
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,631,306
Number of Sequences: 28460
Number of extensions: 88611
Number of successful extensions: 491
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 486
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 488
length of query: 409
length of database: 4,897,600
effective HSP length: 92
effective length of query: 317
effective length of database: 2,279,280
effective search space: 722531760
effective search space used: 722531760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123593.9


