

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
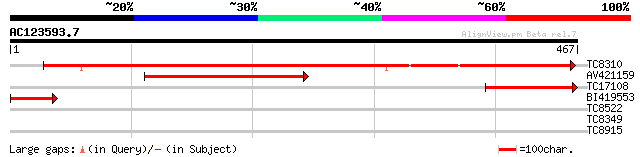
Query= AC123593.7 + phase: 0
(467 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8310 UP|Q9SPM8 (Q9SPM8) Nod factor binding lectin-nucleotide p... 520 e-148
AV421159 231 2e-61
TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, pa... 146 6e-36
BI419553 73 1e-13
TC8522 similar to UP|AAR23736 (AAR23736) At5g46870, partial (76%) 29 1.9
TC8349 28 3.2
TC8915 similar to UP|Q8VYH4 (Q8VYH4) AT5g15260/F8M21_150, partia... 28 3.2
>TC8310 UP|Q9SPM8 (Q9SPM8) Nod factor binding lectin-nucleotide
phosphohydrolase, complete
Length = 1489
Score = 520 bits (1338), Expect = e-148
Identities = 260/443 (58%), Positives = 344/443 (76%), Gaps = 5/443 (1%)
Frame = +1
Query: 29 ILLITFVLYMMPSSSNYDSAGDYALLNRK--LSPDKKSGGSYAVIFDAGSSGSRVHVFHF 86
I L+TFV +MP+ S+ G+ L+NRK L +++ SYAVIFDAGS+GSRVHV++F
Sbjct: 55 ISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNF 234
Query: 87 DQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRI 146
DQNLDL+ + +LE ++ +KPGLS+YA P++AAESL+ LL++AE VVP + TPV++
Sbjct: 235 DQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKL 414
Query: 147 GATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGN 206
GATAGLR LEG+A++ IL+AVRD+L +RS+ +DAV++LDGTQEG+Y WVTINYLLG
Sbjct: 415 GATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGK 594
Query: 207 LGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHS 266
LGK ++KTVGVVDLGGGSVQM YA+S + A AP+V +GEDPY+K++ L+G+KY LYVHS
Sbjct: 595 LGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHS 774
Query: 267 YLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHK 324
YLRYG A RAEI KVAG + NPCIL+G DG Y Y G +K S +SG++LN+C+ +A K
Sbjct: 775 YLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALK 954
Query: 325 ALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPAD 384
ALKVN + C + CTFGGIWNGGGG GQKNLF+ S F+ + + G + N P A +RP D
Sbjct: 955 ALKVN-APCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKIRPVD 1128
Query: 385 FEDAAKQACQTKLENAKSTYPRV-EEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKK 443
+ AAK AC+T LE+AKS YP + E+ ++ Y+C+DLVY YTLLVDGFG+ P+QE+T+ +
Sbjct: 1129LKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANE 1308
Query: 444 VKYDDALVEAAWPLGSAIEAVSS 466
++Y DALVEAAWPLG+AIEA+SS
Sbjct: 1309IEYQDALVEAAWPLGTAIEAISS 1377
>AV421159
Length = 410
Score = 231 bits (589), Expect = 2e-61
Identities = 118/135 (87%), Positives = 127/135 (93%)
Frame = +2
Query: 112 YAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLL 171
YAQ PQQAAESLVSLL+KAE VVPRELRSKT VR+GATAGLRALEGDASD+IL+AVRDLL
Sbjct: 5 YAQNPQQAAESLVSLLDKAESVVPRELRSKTQVRVGATAGLRALEGDASDRILQAVRDLL 184
Query: 172 KHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAI 231
K RSS KS+ADAVTVLDG QEGA+QWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAI
Sbjct: 185 KKRSSLKSEADAVTVLDGVQEGAFQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAI 364
Query: 232 SESEAAMAPQVMDGE 246
SE+EAA AP+V DG+
Sbjct: 365 SETEAAQAPKVTDGQ 409
>TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (16%)
Length = 573
Score = 146 bits (369), Expect = 6e-36
Identities = 67/75 (89%), Positives = 73/75 (97%)
Frame = +1
Query: 393 CQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVE 452
CQTKL +AKSTYP +E+GNLPY+CMDLVYQYTLLVDGFGIYPWQE+TLVKKVKYDDALVE
Sbjct: 16 CQTKLGDAKSTYPHIEDGNLPYICMDLVYQYTLLVDGFGIYPWQEVTLVKKVKYDDALVE 195
Query: 453 AAWPLGSAIEAVSST 467
AAWPLGSAIEAVSS+
Sbjct: 196 AAWPLGSAIEAVSSS 240
>BI419553
Length = 480
Score = 72.8 bits (177), Expect = 1e-13
Identities = 35/39 (89%), Positives = 38/39 (96%)
Frame = +2
Query: 1 MLKRPTRQESLSDKIYRFRGTLLVVSIPILLITFVLYMM 39
MLKRP RQESLSDKIYRFRGTLLV+SIP++LITFVLYMM
Sbjct: 362 MLKRPGRQESLSDKIYRFRGTLLVISIPLVLITFVLYMM 478
>TC8522 similar to UP|AAR23736 (AAR23736) At5g46870, partial (76%)
Length = 1069
Score = 28.9 bits (63), Expect = 1.9
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = -3
Query: 311 SGASLNECKSVAHKALKVNESTCTHMKCTF 340
+GA L EC S+ AL +E TC + TF
Sbjct: 902 AGAGLEECTSLVAAALGDSERTCAYWSSTF 813
>TC8349
Length = 749
Score = 28.1 bits (61), Expect = 3.2
Identities = 16/37 (43%), Positives = 22/37 (59%)
Frame = +2
Query: 278 EILKVAGDAENPCILSGSDGTYKYGGKSFKASSSGAS 314
+ L+VAGD N +SGS+G SFKA+S+ S
Sbjct: 203 QTLRVAGDTSN---VSGSNGPVPSRSNSFKAASNSDS 304
>TC8915 similar to UP|Q8VYH4 (Q8VYH4) AT5g15260/F8M21_150, partial (29%)
Length = 575
Score = 28.1 bits (61), Expect = 3.2
Identities = 12/28 (42%), Positives = 21/28 (74%)
Frame = -2
Query: 27 IPILLITFVLYMMPSSSNYDSAGDYALL 54
+P+ +F+L + P+SS++DS+GD LL
Sbjct: 382 LPMSQESFIL*LYPTSSSFDSSGDPELL 299
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,928,263
Number of Sequences: 28460
Number of extensions: 86447
Number of successful extensions: 455
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 445
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 450
length of query: 467
length of database: 4,897,600
effective HSP length: 94
effective length of query: 373
effective length of database: 2,222,360
effective search space: 828940280
effective search space used: 828940280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC123593.7


