

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123571.8 - phase: 0
(522 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
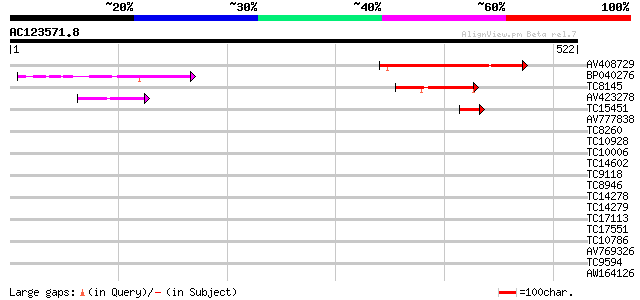
Score E
Sequences producing significant alignments: (bits) Value
AV408729 213 8e-56
BP040276 95 3e-20
TC8145 weakly similar to UP|Q94A38 (Q94A38) AT5g46250/MPL12_3, p... 60 9e-10
AV423278 55 2e-08
TC15451 42 3e-04
AV777838 37 0.008
TC8260 similar to UP|Q94ES8 (Q94ES8) Nodule extensin (Fragment),... 37 0.008
TC10928 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imp... 37 0.010
TC10006 similar to UP|P93396 (P93396) Transformer-SR ribonucleop... 37 0.010
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 37 0.010
TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding l... 36 0.013
TC8946 homologue to GB|AAM66972.1|21617922|AY088650 pyridoxine b... 36 0.013
TC14278 homologue to UP|O65760 (O65760) Extensin (Fragment), par... 36 0.018
TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycop... 36 0.018
TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protei... 35 0.039
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 35 0.039
TC10786 weakly similar to UP|Q9FMM3 (Q9FMM3) Gb|AAC80623.1, part... 35 0.039
AV769326 33 0.087
TC9594 weakly similar to GB|AAM19966.1|20466089|AY098956 At2g326... 33 0.087
AW164126 33 0.11
>AV408729
Length = 433
Score = 213 bits (541), Expect = 8e-56
Identities = 106/139 (76%), Positives = 120/139 (86%), Gaps = 3/139 (2%)
Frame = +3
Query: 341 FVPPMP---LYYAGPDPQLHSKIVNQIDYYFSNENLVKDIFLRKNMDAQGWVPITLIAGF 397
FVPP+P +++AG DPQLH+KIVNQIDYYFSNENLVKD FLR+NMD QGWVPI LIAGF
Sbjct: 3 FVPPVPHHAMFFAGSDPQLHTKIVNQIDYYFSNENLVKDTFLRQNMDDQGWVPIKLIAGF 182
Query: 398 KKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIMPSPVQFPNVTSPEVLNQDM 457
KKVM LTDNIQLI+DA++TS VVEVQGDKIRR+NDW +WIMPSP QFPNVTS VLN D
Sbjct: 183 KKVMHLTDNIQLILDAVQTSYVVEVQGDKIRRRNDWMRWIMPSP-QFPNVTSHGVLNHDK 359
Query: 458 LAEKMRNIALETTIYDGAG 476
LAE++ NI+LETT YD G
Sbjct: 360 LAEQLHNISLETTNYDIPG 416
>BP040276
Length = 556
Score = 94.7 bits (234), Expect = 3e-20
Identities = 67/166 (40%), Positives = 86/166 (51%), Gaps = 2/166 (1%)
Frame = +1
Query: 8 SSNHSPDNLQSRRLPPWNQVVRGESESIAAVPAVSLSEESFPIVTAPVDDSTSAEVISDN 67
S NHSP PW+Q+V + ++ PAVS+S+ S + +AP +++
Sbjct: 133 SPNHSPSP------SPWSQIVSAAAPP-SSPPAVSVSDSS-AVNSAPAEET--------- 261
Query: 68 ADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKS--ESSKGLL 125
E N GKRPAWN+ S S VM A SWPALS SA KS ES KG +
Sbjct: 262 -----ENNANGGKRPAWNKPSNAASSS----VMGADSWPALSKSAHSPAKSLPESVKGSV 414
Query: 126 DGSSVSPWQGMESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSSN 171
D SS SP + + S QRQV DN VNN PT+QK K ++SN
Sbjct: 415 DSSSPSPNLQGTGSATPSPQRQVRDNAGVNNTLPTQQKPYKRSNSN 552
>TC8145 weakly similar to UP|Q94A38 (Q94A38) AT5g46250/MPL12_3, partial
(49%)
Length = 1671
Score = 60.1 bits (144), Expect = 9e-10
Identities = 34/82 (41%), Positives = 55/82 (66%), Gaps = 6/82 (7%)
Frame = +1
Query: 356 LHSKIVNQIDYYFSNENLVKDI----FLRKNMDAQGWVPITLIAGFKKVMDLTDNIQLII 411
L KI+ Q++YYFS+ENL D FL+KN +G+VP ++IA F+K+ LT + +I+
Sbjct: 340 LKLKIIRQVEYYFSDENLPNDKYLLGFLKKN--REGFVPTSVIASFRKMKRLTRDHSIIV 513
Query: 412 DAIRTSSVVEVQGD--KIRRQN 431
A++ SS++ V GD +++R N
Sbjct: 514 AALKESSLLVVSGDGRRVKRLN 579
>AV423278
Length = 389
Score = 55.5 bits (132), Expect = 2e-08
Identities = 29/66 (43%), Positives = 39/66 (58%)
Frame = +2
Query: 63 VISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSESSK 122
+I D++D +N G K+PAW + V+EV PVM A SWPALS++A+ S K S
Sbjct: 185 LIHDSSDADKAKNAGAPKKPAWKMPPSD--VAEVSPVMGAVSWPALSEAAKLSVKPTSDS 358
Query: 123 GLLDGS 128
DGS
Sbjct: 359 STTDGS 376
>TC15451
Length = 662
Score = 41.6 bits (96), Expect = 3e-04
Identities = 16/23 (69%), Positives = 21/23 (90%)
Frame = +1
Query: 415 RTSSVVEVQGDKIRRQNDWEKWI 437
RTS+VVEVQGD +RR+NDW +W+
Sbjct: 1 RTSTVVEVQGDLLRRRNDWMRWL 69
>AV777838
Length = 565
Score = 37.0 bits (84), Expect = 0.008
Identities = 32/119 (26%), Positives = 47/119 (38%), Gaps = 1/119 (0%)
Frame = +1
Query: 169 SSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGF 228
+S+ GHT + P S +++ S +SSS + S R T + +P + S
Sbjct: 166 ASSRELAAGHTTITGPPSSSSSSSSSSSSSSSSSSSMR-ATAAGHTTITAPPSSSSSSMR 342
Query: 229 VPSDHPQQRNSFRHR-NGGPHQRGDGSHHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVS 286
+ H +S H G P GSHHHH + RR + D PP S
Sbjct: 343 ATAGHTITSSSSSHEPKGQPPWPPRGSHHHHENLGPPPSKPDLRRTAHDLDKPTPPLCS 519
>TC8260 similar to UP|Q94ES8 (Q94ES8) Nodule extensin (Fragment), partial
(79%)
Length = 1017
Score = 37.0 bits (84), Expect = 0.008
Identities = 25/91 (27%), Positives = 36/91 (39%), Gaps = 13/91 (14%)
Frame = +3
Query: 282 PPRVSPRIIRPSLPPNSAPFIHP------PPLRPFGGHMGF-------HELAAPVVLFAG 328
PP + P PP+ P HP PP P H + H P ++
Sbjct: 276 PPHKPYKYPSPPPPPHKYPHPHPHPVYHSPPPPPPKKHYKYSSPPPPVHTYPHPHPVYHS 455
Query: 329 PPPPIDSLRGVPFVPPMPLYYAGPDPQLHSK 359
PPPP+ + + P P+Y++ P P H K
Sbjct: 456 PPPPVHT-----YPHPHPVYHSPPPPPPHEK 533
Score = 32.3 bits (72), Expect = 0.19
Identities = 19/60 (31%), Positives = 27/60 (44%), Gaps = 6/60 (10%)
Frame = +3
Query: 304 PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV------PFVPPMPLYYAGPDPQLH 357
PPP +P+ +H PV PPPP + V P+ P P+Y++ P P H
Sbjct: 126 PPPPKPYS----YHSPPPPV---HSPPPPYEKPHPVYHSPPPPYEKPHPVYHSPPPPPPH 284
Score = 31.2 bits (69), Expect = 0.43
Identities = 29/96 (30%), Positives = 37/96 (38%), Gaps = 13/96 (13%)
Frame = +3
Query: 279 MHVPPRVSPRIIRPSLP----PNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPID 334
+H P P P P P+ P H PP P H ++ A+P PPPP+
Sbjct: 417 VHTYPHPHPVYHSPPPPVHTYPHPHPVYHSPPPPP--PHEKPYKYASP------PPPPVH 572
Query: 335 SL-RGVPFVPPMPL--------YYAGPDPQLHSKIV 361
+ V PP P+ YY P P H IV
Sbjct: 573 TYPHPVYHSPPPPVHSPPPPHYYYKSPPPPYHH*IV 680
Score = 29.6 bits (65), Expect = 1.3
Identities = 24/78 (30%), Positives = 27/78 (33%), Gaps = 5/78 (6%)
Frame = +3
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPV-----VLFAGPPPPIDSL 336
PP P PP +P PPP H +H P V + PPPP
Sbjct: 126 PPPPKPYSYHSPPPPVHSP---PPPYEK--PHPVYHSPPPPYEKPHPVYHSPPPPPPHKP 290
Query: 337 RGVPFVPPMPLYYAGPDP 354
P PP P Y P P
Sbjct: 291 YKYPSPPPPPHKYPHPHP 344
>TC10928 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(29%)
Length = 590
Score = 36.6 bits (83), Expect = 0.010
Identities = 38/133 (28%), Positives = 50/133 (37%), Gaps = 13/133 (9%)
Frame = -3
Query: 235 QQRNSFRHRNGGPHQRGDGSHH-------HHNYGNRRDQDWNSRRNYNGRDMHVPPRVSP 287
+Q N+ H + SHH HHN + Q W ++N + + P VS
Sbjct: 543 KQGNALTHSHYT*DTSQSDSHHSTPTCNFHHN--PNKAQTWPHHTHHNHISLPLSPHVSH 370
Query: 288 RIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI------DSLRGVPF 341
P+ P +SAP PP RP AAP PPPP ++ G P
Sbjct: 369 SRSDPTDPTHSAP--PTPPTRPNTSPSQTPSAAAPPP--PPPPPPAAPSPLSETRSGAPR 202
Query: 342 VPPMPLYYAGPDP 354
P YA P P
Sbjct: 201 HARPPPLYAPPAP 163
>TC10006 similar to UP|P93396 (P93396) Transformer-SR ribonucleoprotein
(Fragment), partial (30%)
Length = 453
Score = 36.6 bits (83), Expect = 0.010
Identities = 28/110 (25%), Positives = 50/110 (45%), Gaps = 8/110 (7%)
Frame = +3
Query: 153 NVNNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSP 212
N N + S+ +S++ S + +P A + S + S R ++SPR ++SP
Sbjct: 36 NPNPSWSSHSSSLFPDSTSMSDSPVRRNSRSPSPWRAQSRSRSRSPRPKSRSPRPKSRSP 215
Query: 213 RDHAQSPRDHTQ------RSGFVPSDHPQQRNSF--RHRNGGPHQRGDGS 254
R ++SPR ++ RS + P+ R+ R R+ P R G+
Sbjct: 216 RAKSRSPRPRSRSPRPRSRSRSLEKQRPRSRSRSRGRSRSRSPEDRNGGN 365
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 36.6 bits (83), Expect = 0.010
Identities = 29/117 (24%), Positives = 46/117 (38%)
Frame = -1
Query: 180 QQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNS 239
Q+S PQ SH++ +T+SP T P H SP + + P +S
Sbjct: 698 QKSPPQNHRKTAASHSTL---YTKSPHSSTHPPSPHTPSPSKPPAQHN--TASQPPHSSS 534
Query: 240 FRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPRIIRPSLPP 296
RH+ P+Q S HH+ + + G D P + + ++P PP
Sbjct: 533 SRHQTTPPNQNPPVSDLHHHQTKHYPVSHHGAQ-LQGGDSAPPLQEAASSVQPLQPP 366
>TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding
like-protein, partial (16%)
Length = 779
Score = 36.2 bits (82), Expect = 0.013
Identities = 23/84 (27%), Positives = 31/84 (36%), Gaps = 4/84 (4%)
Frame = +2
Query: 199 RDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSH--- 255
RDH +S + H H RD R H ++ + +R G H D H
Sbjct: 71 RDHKRSRKKHRSHRSSHTSRDRDRDGRKH--KRRHSSSKDKYSYR-GAKHGISDDEHQHS 241
Query: 256 -HHHNYGNRRDQDWNSRRNYNGRD 278
HHH Y + D+ S R D
Sbjct: 242 SHHHKYDSSSDEKHRSSRRRQRED 313
>TC8946 homologue to GB|AAM66972.1|21617922|AY088650 pyridoxine
biosynthesis protein-like {Arabidopsis thaliana;} ,
partial (45%)
Length = 503
Score = 36.2 bits (82), Expect = 0.013
Identities = 37/134 (27%), Positives = 46/134 (33%), Gaps = 7/134 (5%)
Frame = +3
Query: 178 HTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQR 237
H Q P + + S R H + R T R ++ R H R+ P HP+ R
Sbjct: 129 HRDQEIPLLRQSRPRSDAPRRRHHGRRQRP-TSPHRRRSRRLRRHGPRA--CPRRHPRPR 299
Query: 238 NSFRHRNGGPHQRGDGSHHHHNYGN------RRDQDWNS-RRNYNGRDMHVPPRVSPRII 290
H HQ S HH +G RR D S RR R+ P
Sbjct: 300 RRRSHERPSAHQGNQTSRHHPRHGQSPHRPFRRSPDPRSHRRRLRRRER------GPHSR 461
Query: 291 RPSLPPNSAPFIHP 304
R P A F HP
Sbjct: 462 RRG*PHQQAQFPHP 503
>TC14278 homologue to UP|O65760 (O65760) Extensin (Fragment), partial (28%)
Length = 619
Score = 35.8 bits (81), Expect = 0.018
Identities = 22/65 (33%), Positives = 28/65 (42%), Gaps = 1/65 (1%)
Frame = +2
Query: 294 LPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLY-YAGP 352
LPP + PPP P AP ++ PPPP S PP P+Y YA P
Sbjct: 17 LPPPPYHYTSPPPPSPS---------PAPTYIYKSPPPPTKS-------PPPPVYIYASP 148
Query: 353 DPQLH 357
P ++
Sbjct: 149 PPPIY 163
Score = 27.3 bits (59), Expect = 6.2
Identities = 18/51 (35%), Positives = 21/51 (40%)
Frame = +2
Query: 283 PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI 333
P +P I S PP P PPP PV ++A PPPPI
Sbjct: 62 PSPAPTYIYKSPPP---PTKSPPP---------------PVYIYASPPPPI 160
>TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (57%)
Length = 941
Score = 35.8 bits (81), Expect = 0.018
Identities = 23/70 (32%), Positives = 29/70 (40%)
Frame = +1
Query: 285 VSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPP 344
VSP PS PP + PPP P AP ++ PPPP+ +PP
Sbjct: 379 VSPPPPSPS-PPPPYHYASPPPPSPS---------PAPTYIYKSPPPPVK-------LPP 507
Query: 345 MPLYYAGPDP 354
P +Y P P
Sbjct: 508 PPYHYTSPPP 537
Score = 35.8 bits (81), Expect = 0.018
Identities = 22/65 (33%), Positives = 28/65 (42%), Gaps = 1/65 (1%)
Frame = +1
Query: 294 LPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLY-YAGP 352
LPP + PPP P AP ++ PPPP S PP P+Y YA P
Sbjct: 499 LPPPPYHYTSPPPPSPS---------PAPTYIYKSPPPPTKS-------PPPPVYIYASP 630
Query: 353 DPQLH 357
P ++
Sbjct: 631 PPPIY 645
Score = 34.7 bits (78), Expect = 0.039
Identities = 19/55 (34%), Positives = 21/55 (37%)
Frame = +1
Query: 304 PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHS 358
PPP P + P + PPPP S PP P YY P P HS
Sbjct: 22 PPPPPPVPKPYYYKSPPPPPYYYKSPPPPSPS-------PPPPYYYQSPPPPSHS 165
Score = 31.2 bits (69), Expect = 0.43
Identities = 23/80 (28%), Positives = 28/80 (34%), Gaps = 7/80 (8%)
Frame = +1
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPV-------VLFAGPPPPID 334
PP V S PP + PPP P +++ P + PPPP
Sbjct: 31 PPPVPKPYYYKSPPPPPYYYKSPPPPSPSPPPPYYYQSPPPPSHSPPPPYYYKSPPPPSP 210
Query: 335 SLRGVPFVPPMPLYYAGPDP 354
S PP P YY P P
Sbjct: 211 S-------PPPPYYYKSPPP 249
Score = 30.0 bits (66), Expect = 0.96
Identities = 23/74 (31%), Positives = 26/74 (35%), Gaps = 1/74 (1%)
Frame = +1
Query: 282 PPRVSPRIIRPSLPPNSAPFIH-PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVP 340
PP SP P + P H PPP + P + PPPP S
Sbjct: 97 PPPPSPSPPPPYYYQSPPPPSHSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPS----- 261
Query: 341 FVPPMPLYYAGPDP 354
PP P YY P P
Sbjct: 262 --PPPPYYYKSPPP 297
Score = 27.3 bits (59), Expect = 6.2
Identities = 18/51 (35%), Positives = 21/51 (40%)
Frame = +1
Query: 283 PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI 333
P +P I S PP P PPP PV ++A PPPPI
Sbjct: 544 PSPAPTYIYKSPPP---PTKSPPP---------------PVYIYASPPPPI 642
>TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protein, partial
(25%)
Length = 1077
Score = 34.7 bits (78), Expect = 0.039
Identities = 25/77 (32%), Positives = 29/77 (37%), Gaps = 6/77 (7%)
Frame = +1
Query: 282 PPRVSPRIIRPSLPPNSAP------FIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDS 335
PP P I P PP S P F PP +P F + P + PPPP
Sbjct: 595 PPSPPPLIPNPFQPPPSPPPFIPNPFQPPPSKQPPSPLFPFPPIVIPGLTPPPPPPPPP- 771
Query: 336 LRGVPFVPPMPLYYAGP 352
PF PP P + P
Sbjct: 772 ----PFFPPFPPLFPPP 810
Score = 29.3 bits (64), Expect = 1.6
Identities = 28/86 (32%), Positives = 31/86 (35%), Gaps = 7/86 (8%)
Frame = +1
Query: 278 DMHVPPRVSPRIIRPSLPPNSAPFI----HPPPLRPFGGHMGFH---ELAAPVVLFAGPP 330
D PP P P PP+ P I PPP P F P LF PP
Sbjct: 544 DPFQPPSPPPLFPNPFQPPSPPPLIPNPFQPPPSPPPFIPNPFQPPPSKQPPSPLFPFPP 723
Query: 331 PPIDSLRGVPFVPPMPLYYAGPDPQL 356
I L P PP P ++ P P L
Sbjct: 724 IVIPGLTPPPPPPPPPPFFP-PFPPL 798
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 34.7 bits (78), Expect = 0.039
Identities = 31/110 (28%), Positives = 41/110 (37%), Gaps = 7/110 (6%)
Frame = +1
Query: 255 HHHHNYGNRRDQDWNSRRN-------YNGRDMHVPPRVSPRIIRPSLPPNSAPFIHPPPL 307
HHHH++ +RR + + ++ R H P + P P LP P + PPPL
Sbjct: 70 HHHHHHHHRRQTTATTSHHHFLPP*RHHSRLPHPRPTLFPPFPLPRLP----PLVPPPPL 237
Query: 308 RPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLH 357
P +L PPPP PP PL P P H
Sbjct: 238 --------------PRLLRPPPPPP----------PPPPL---RPRPHRH 306
>TC10786 weakly similar to UP|Q9FMM3 (Q9FMM3) Gb|AAC80623.1, partial (9%)
Length = 756
Score = 34.7 bits (78), Expect = 0.039
Identities = 34/153 (22%), Positives = 57/153 (37%), Gaps = 6/153 (3%)
Frame = +3
Query: 136 MESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHT 195
+ES P S VG+NV T +++K ++ + GH Q V +
Sbjct: 222 VESKPGSG---NVGNNVISIPPYGTHSETVK---TSGNGEDGHDVQKRKDVFRPSMLDSE 383
Query: 196 SSSRDHTQSPRDHTQSP------RDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQ 249
S RD + T+S RD + D + + S + R H+
Sbjct: 384 SGRRDRWRDEERETKSATRKDRWRDGDKDLGDSRKVDRWQDSLPSKNLGEARRGASDSHR 563
Query: 250 RGDGSHHHHNYGNRRDQDWNSRRNYNGRDMHVP 282
R D + N+ RR+ WN+R + ++ P
Sbjct: 564 RNDSGNRETNFDQRRESKWNTRWGPDNKEPEGP 662
>AV769326
Length = 262
Score = 33.5 bits (75), Expect = 0.087
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 2/53 (3%)
Frame = +3
Query: 290 IRPSLPPNSAPF--IHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVP 340
+R + PP P + PPP RP H L P++ PPPPI +LR P
Sbjct: 12 LRGAFPPRRPPLPALLPPPRRPRPLHPPILTLLPPIL---PPPPPIRTLRASP 161
>TC9594 weakly similar to GB|AAM19966.1|20466089|AY098956
At2g32600/T26B15.16 {Arabidopsis thaliana;}, partial
(9%)
Length = 502
Score = 33.5 bits (75), Expect = 0.087
Identities = 12/22 (54%), Positives = 13/22 (58%)
Frame = +1
Query: 248 HQRGDGSHHHHNYGNRRDQDWN 269
H R G HHHHN NR D W+
Sbjct: 202 HHRFGGHHHHHNISNRLDARWH 267
>AW164126
Length = 387
Score = 33.1 bits (74), Expect = 0.11
Identities = 25/88 (28%), Positives = 34/88 (38%)
Frame = +2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP SP+ PP +P PPP + + P + PPPP
Sbjct: 149 PPSPSPKPYYYKSPPPPSP--SPPPPYYYKSPPPPSPVPHPPYYYKSPPPPSP------- 301
Query: 342 VPPMPLYYAGPDPQLHSKIVNQIDYYFS 369
+P P YY+ P P + S YY+S
Sbjct: 302 IPHPPYYYSSPPPPVKSP-PPPTPYYYS 382
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.130 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,977,815
Number of Sequences: 28460
Number of extensions: 184403
Number of successful extensions: 1578
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 1390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1515
length of query: 522
length of database: 4,897,600
effective HSP length: 94
effective length of query: 428
effective length of database: 2,222,360
effective search space: 951170080
effective search space used: 951170080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC123571.8


