

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123570.3 + phase: 0 /pseudo
(1023 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
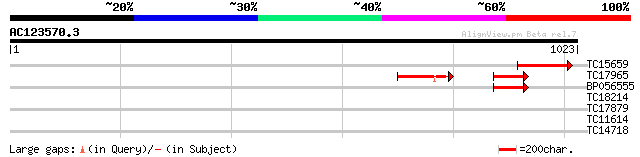
Score E
Sequences producing significant alignments: (bits) Value
TC15659 116 2e-26
TC17965 96 3e-20
BP056555 94 1e-19
TC18214 similar to UP|SEC5_ARATH (Q8S3U9) Exocyst complex compon... 30 1.5
TC17879 similar to AAQ56782 (AAQ56782) At2g27460, partial (32%) 30 2.5
TC11614 similar to UP|Q9FHH4 (Q9FHH4) Diacylglycerol kinase-like... 29 4.3
TC14718 similar to UP|NCPR_CATRO (Q05001) NADPH--cytochrome P450... 28 9.7
>TC15659
Length = 574
Score = 116 bits (290), Expect = 2e-26
Identities = 67/101 (66%), Positives = 77/101 (75%), Gaps = 2/101 (1%)
Frame = +1
Query: 916 DKDILYGLLLVLSGMLTEKN--ESLLFKAIQIKNDVNG*FLFLPILQLVRESCIQCLVAL 973
DKDILY LLLVLSG+LTEKN E++ A I N + + P LVRE+ +QCLVAL
Sbjct: 1 DKDILYVLLLVLSGILTEKNGQEAVTENAHIIINCLIK-LVDYPHKTLVRETAVQCLVAL 177
Query: 974 SKLPHVRIYPLRTQVLEAISKCLDDTKRSVRNEAVKCRQAW 1014
S LPHVRIYP+RTQVL AISKCLDD KR+VR EAVKC+QAW
Sbjct: 178 SVLPHVRIYPMRTQVLRAISKCLDDRKRAVRLEAVKCQQAW 300
>TC17965
Length = 575
Score = 95.9 bits (237), Expect = 3e-20
Identities = 49/63 (77%), Positives = 57/63 (89%)
Frame = +1
Query: 873 RYLLLRAFARVMSVTPLIVILNDAKELISVLLDCLSMLTEDIQDKDILYGLLLVLSGMLT 932
R L RAFA ++S TPLIVI+++AK+LI VLL+CLSMLTEDIQDKDILYGLLLVLSG+LT
Sbjct: 214 RSFLYRAFAHIISDTPLIVIVSEAKKLIPVLLNCLSMLTEDIQDKDILYGLLLVLSGILT 393
Query: 933 EKN 935
EKN
Sbjct: 394 EKN 402
Score = 72.8 bits (177), Expect = 3e-13
Identities = 43/105 (40%), Positives = 66/105 (61%), Gaps = 4/105 (3%)
Frame = +1
Query: 701 AEKSDELTLEEALHIIFNTKICFSSDNMLQICDGSINRNEIVLTDVCLGMTNDRLLQTNA 760
AE S+ LTLEEAL++IF+TKI S ++LQ C+G+ N +E+VLT+ CLG+ NDRLLQ++
Sbjct: 1 AENSNNLTLEEALNVIFSTKIWVCSTDVLQGCNGADNGSEMVLTE*CLGIANDRLLQSSN 180
Query: 761 VCGL----SWIGKGLLLRGHEKIKDITKILTECLISDRNSSLPLI 801
C L W + L R I T ++ ++S+ +P++
Sbjct: 181 QCHLWALMDW-KRSFLYRAFAHIISDTPLIV--IVSEAKKLIPVL 306
>BP056555
Length = 428
Score = 93.6 bits (231), Expect = 1e-19
Identities = 48/63 (76%), Positives = 56/63 (88%)
Frame = -3
Query: 873 RYLLLRAFARVMSVTPLIVILNDAKELISVLLDCLSMLTEDIQDKDILYGLLLVLSGMLT 932
R L RAFA ++S TPLIVI+++AK+LI VLL+CLSMLTEDI DKDILYGLLLVLSG+LT
Sbjct: 417 RSFLYRAFAHIISDTPLIVIVSEAKKLIPVLLNCLSMLTEDIPDKDILYGLLLVLSGILT 238
Query: 933 EKN 935
EKN
Sbjct: 237 EKN 229
>TC18214 similar to UP|SEC5_ARATH (Q8S3U9) Exocyst complex component Sec5,
partial (20%)
Length = 656
Score = 30.4 bits (67), Expect = 1.5
Identities = 11/37 (29%), Positives = 22/37 (58%)
Frame = -1
Query: 819 KCAADAFHVLMSDAEDCLNRKFHATMRPLYKQRFFSS 855
KC+ + M +A++C+N+ FH + ++RF +S
Sbjct: 296 KCSKILTFIFMKNADECVNQAFHKNAKCFIQERFTTS 186
>TC17879 similar to AAQ56782 (AAQ56782) At2g27460, partial (32%)
Length = 953
Score = 29.6 bits (65), Expect = 2.5
Identities = 20/53 (37%), Positives = 29/53 (53%), Gaps = 3/53 (5%)
Frame = -2
Query: 392 KGLQILAMFNLDVFPIPKSTFENILKKFMSIII---EDFNKTILWNSTLKSLF 441
KGLQ+ + D+ TF +IL KF+SII+ +D + NSTL + F
Sbjct: 205 KGLQLRTLRECDIKLRSYGTFISILNKFISIIVLSRKDH*PFLKQNSTLPNSF 47
>TC11614 similar to UP|Q9FHH4 (Q9FHH4) Diacylglycerol kinase-like protein,
partial (24%)
Length = 634
Score = 28.9 bits (63), Expect = 4.3
Identities = 15/47 (31%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Frame = -3
Query: 978 HVRIYPLRTQVLEAISKCLDDTKRSVR-NEAVKCRQAWLVFQIFQEP 1023
H+ ++P T ++ S CL R+ + N +C WL + FQEP
Sbjct: 278 HIGVFPASTSEFKSNS*CLS*MLRTDQFNHDKRCMPCWL*AKYFQEP 138
>TC14718 similar to UP|NCPR_CATRO (Q05001) NADPH--cytochrome P450 reductase
(CPR) (P450R) , partial (36%)
Length = 1163
Score = 27.7 bits (60), Expect = 9.7
Identities = 16/55 (29%), Positives = 28/55 (50%)
Frame = -3
Query: 661 HVPNIRGILHLFIITLLKGVVPVAQALGSMVNKLISKSNGAEKSDELTLEEALHI 715
HVPNIR +LH F++ + G ++L N + K +K +L + +H+
Sbjct: 606 HVPNIRSLLHHFMLHIFLG-----RSLP*EGNNELRKRTTVDKVVQLIFIDVVHL 457
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,845,024
Number of Sequences: 28460
Number of extensions: 261253
Number of successful extensions: 1422
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 1407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1421
length of query: 1023
length of database: 4,897,600
effective HSP length: 99
effective length of query: 924
effective length of database: 2,080,060
effective search space: 1921975440
effective search space used: 1921975440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC123570.3


