

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
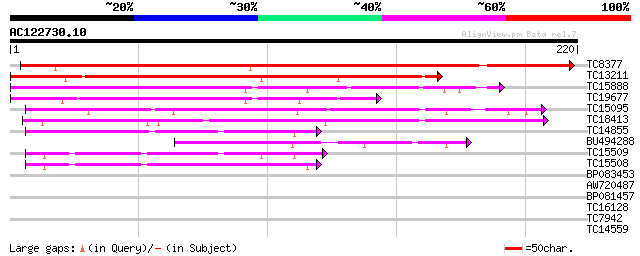
Query= AC122730.10 - phase: 0
(220 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8377 similar to UP|Q9M3Z7 (Q9M3Z7) Alpha-fucosidase , partial... 260 1e-70
TC13211 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibito... 140 2e-34
TC15888 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibito... 121 8e-29
TC19677 weakly similar to UP|O82711 (O82711) Profucosidase precu... 108 9e-25
TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein... 90 3e-19
TC18413 similar to UP|P93378 (P93378) Tumor-related protein, par... 69 5e-13
TC14855 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precurs... 54 2e-08
BU494288 53 4e-08
TC15509 weakly similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor ... 49 5e-07
TC15508 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precurs... 48 1e-06
BP083453 27 3.6
AW720487 26 4.7
BP081457 26 6.2
TC16128 homologue to UP|PSE1_ARATH (Q9S831) Photosystem I reacti... 26 6.2
TC7942 similar to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragmen... 25 8.0
TC14559 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associat... 25 8.0
>TC8377 similar to UP|Q9M3Z7 (Q9M3Z7) Alpha-fucosidase , partial (83%)
Length = 1014
Score = 260 bits (665), Expect = 1e-70
Identities = 130/217 (59%), Positives = 167/217 (76%), Gaps = 2/217 (0%)
Frame = +1
Query: 5 LSLTLSFLLFVF-TTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLG 63
+ L+L FL F F +T L AFS++ VEQVLD+NG+ IFPGG+YYI PAIRGP GGG+RLG
Sbjct: 40 IPLSLCFLFFAFISTKLPTAFSSNDVEQVLDVNGDSIFPGGRYYIRPAIRGPPGGGVRLG 219
Query: 64 KSSNSDCEVTVVQDYNEVINGVPVKFSIP-EISPGIIFTGTPIDIEFTKKPNCVESSKWL 122
K+ +SDC VT +Q+Y+EV NG+PV+F I E+S GIIFTGT +DIEFT KP+CVES KW+
Sbjct: 220 KTGDSDCPVTALQEYSEVKNGIPVRFRIAAEVSTGIIFTGTELDIEFTVKPHCVESPKWI 399
Query: 123 IFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYCVKDSPTCLDIGRAH 182
+FVD+ I KACVGIGG +++PG +T SGTFNI+K+ +GFGY+L +C+K SPTCLDIGR
Sbjct: 400 VFVDNEIGKACVGIGGAKDHPGKQTFSGTFNIQKNSAGFGYKLVFCIKGSPTCLDIGRYD 579
Query: 183 EEVEDEGGSRLHLTHQVAFAVVFVDAASYEAGIIKSV 219
EGG RL+LT AF +VFV ++ I+SV
Sbjct: 580 ---NGEGGRRLNLTEHEAFDLVFVPVGVADSAGIRSV 681
>TC13211 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibitor 20,
partial (44%)
Length = 558
Score = 140 bits (352), Expect = 2e-34
Identities = 81/172 (47%), Positives = 108/172 (62%), Gaps = 4/172 (2%)
Frame = +1
Query: 1 MKHVLSLTLSFLLFVFTTNL-SLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGG 59
MK + L+L F LF T L LAFS D EQV D GNP+ PG +Y I+PA+RG GG
Sbjct: 19 MKPTILLSLCFFLFALTPYLFPLAFSQDP-EQVKDAYGNPLVPGRRYSIVPALRGSRSGG 195
Query: 60 LRLGKSSNSDCEVTVVQDYNEVINGVPVKFSIPEISP--GIIFTGTPIDIEFTKKPNCVE 117
+ K+ N C +TVVQ+Y E +G PV FS+ I+P G I TGT +DI F +KP C +
Sbjct: 196 VSPDKTGNQTCPLTVVQEYFECRDGKPVIFSVSGITPGTGTILTGTHLDIAFVEKPECAD 375
Query: 118 SSKWLIFVD-SVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYC 168
+SKW++ + + A V IGG E++PG L GTFNI+K + G GY+L +C
Sbjct: 376 TSKWVVVGNLTDYPGAPVSIGGVEDHPGQHILDGTFNIQKLDCG-GYKLVFC 528
>TC15888 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibitor 20,
partial (27%)
Length = 611
Score = 121 bits (304), Expect = 8e-29
Identities = 82/197 (41%), Positives = 113/197 (56%), Gaps = 5/197 (2%)
Frame = +3
Query: 1 MKHVLSLTLSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGL 60
MK + LTLS LLF FTT + + + + V D NG P+ G+YYI PA+RGP GGG+
Sbjct: 45 MKPTMLLTLSVLLFAFTTYIIPSAFSQVPDLVKDTNGIPLRSDGRYYIRPALRGPGGGGV 224
Query: 61 RLGKSSNSDCEVTVVQDYNEVINGVPVKFS-IPEISPGIIFTGTPIDIEFTKKPN-CVES 118
G++ N C VTV+Q +EV NG VKF P I GII+ G P+ I F C +S
Sbjct: 225 SPGETGNQTCPVTVLQHKSEVENGNGVKFEHYPNI--GIIYEGVPLTISFGDIYGFCADS 398
Query: 119 SKWLIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGY--CVKDSP-TC 175
+KW++ D K VGIGG E++PG +SG F+I+K++ Y+L + SP
Sbjct: 399 TKWVVVSDDFPGK-WVGIGGAEDHPGKEIVSGLFDIQKYDD-VAYKLVFTPVTGGSPGLS 572
Query: 176 LDIGRAHEEVEDEGGSR 192
+DIGR +D+ G R
Sbjct: 573 VDIGRR----DDKNGRR 611
>TC19677 weakly similar to UP|O82711 (O82711) Profucosidase precursor ,
partial (27%)
Length = 471
Score = 108 bits (269), Expect = 9e-25
Identities = 69/148 (46%), Positives = 86/148 (57%), Gaps = 4/148 (2%)
Frame = +3
Query: 1 MKHVLSLTLSFLLFVFTTN--LSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGG 58
MK + LTLS LLF F+T + LAFS + V DING P+ GKYY+LPAI GP GG
Sbjct: 36 MKPTILLTLSVLLFAFSTTYIIPLAFSQ-VPDLVKDINGIPLSSDGKYYVLPAIWGPTGG 212
Query: 59 GLRLGKSSNSDCEVTVVQDYNEVINGVPVKFS-IPEISPGIIFTGTPIDIEFTK-KPNCV 116
G+ + N C VTV+Q Y EV NG VKF P I GII+ G P+ I F C
Sbjct: 213 GVSPRVTGNQTCPVTVLQSYYEVENGKGVKFEHFPNI--GIIYEGVPLKIYFANIYGYCA 386
Query: 117 ESSKWLIFVDSVIQKACVGIGGPENYPG 144
ES++W++ D VGIGG ++PG
Sbjct: 387 ESTEWVVVSDD-FPTPWVGIGGAADHPG 467
>TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein
(Fragment), partial (42%)
Length = 979
Score = 89.7 bits (221), Expect = 3e-19
Identities = 71/212 (33%), Positives = 105/212 (49%), Gaps = 10/212 (4%)
Frame = +3
Query: 7 LTLSFLLFVFTTNLSLAFSNDAV-EQVLDINGNPIFPGGKYYILPAIRGPLGGGLRL--- 62
L S + F FTT + ++ A E VLDI+G + G KYYILP +RG GGGL L
Sbjct: 201 LAFSIICFAFTTEFLIGIASAAAPEPVLDISGQKLRTGVKYYILPVLRGK-GGGLTLTSS 377
Query: 63 GKSSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFT-KKPNCVESSKW 121
G +N+ C + VVQ+ EV+ G V F+ G+I T T ++I+ + C ++ W
Sbjct: 378 GNINNNTCPLHVVQEKLEVLKGQSVTFTPYNAKGGVILTSTDLNIKSSLTNTTCAKAPVW 557
Query: 122 LIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYC---VKDSPTCLDI 178
+ + + + GG E PG T+S F IEK + Y +C K C ++
Sbjct: 558 KL-LKELSGVWFLATGGVEGNPGMATISNWFKIEKADK--DYVFSFCPSVCKCQTLCREL 728
Query: 179 GRAHEEVEDEGGSR-LHLTHQV-AFAVVFVDA 208
G + D GG + L L+ QV F ++F A
Sbjct: 729 G-----IYDYGGDKHLSLSDQVPPFRIMFKQA 809
>TC18413 similar to UP|P93378 (P93378) Tumor-related protein, partial (41%)
Length = 740
Score = 69.3 bits (168), Expect = 5e-13
Identities = 66/215 (30%), Positives = 92/215 (42%), Gaps = 11/215 (5%)
Frame = -3
Query: 6 SLTLSF-LLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAI------RGPL-- 56
S+ LSF +LF T L + + EQV+D G + G YYI P RGP
Sbjct: 708 SIYLSFAILFALCTQPFLGKGDASPEQVVDTQGKNVRAGMGYYIRPVPTTPCDGRGPCVV 529
Query: 57 -GGGLRLGKSSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKKPNC 115
G + + +S N C + VV E G V F+ G+I T ++I T + C
Sbjct: 528 GSGFVLIARSPNKTCPLNVV--VVEGFRGQAVTFTPVNPKKGVIRVSTDVNINVTLETAC 355
Query: 116 VESSKW-LIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYCVKDSPT 174
ES+ W L D+ V GG PG T+S F IEK+E Y+L +C T
Sbjct: 354 EESTVWKLDDFDASAGHWFVTTGGVVGNPGKDTISNWFKIEKYEE--DYKLVFCPTVCDT 181
Query: 175 CLDIGRAHEEVEDEGGSRLHLTHQVAFAVVFVDAA 209
C + R D G+R ++ V F +A
Sbjct: 180 CKPLCRNVGVFMDSNGNRRVALTDESYKVRFQPSA 76
>TC14855 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(90%)
Length = 1001
Score = 54.3 bits (129), Expect = 2e-08
Identities = 35/116 (30%), Positives = 54/116 (46%), Gaps = 1/116 (0%)
Frame = +1
Query: 7 LTLSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLGKSS 66
L L FL + L + E V+D GNP+ PG YY P GGL LG++
Sbjct: 64 LPLFFLSLLILNTQPLLAAEPEPEPVVDKQGNPLEPGVGYYAWPLWAD--NGGLTLGRTR 237
Query: 67 NSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEF-TKKPNCVESSKW 121
N C + +++D +++ G P++F + + G I T T + +E C E W
Sbjct: 238 NKTCPLDIIRDPSKI--GTPIEFHVADSDLGYIPTLTDLTVEMPILGSRCQEPKVW 399
>BU494288
Length = 438
Score = 53.1 bits (126), Expect = 4e-08
Identities = 42/124 (33%), Positives = 56/124 (44%), Gaps = 9/124 (7%)
Frame = +3
Query: 65 SSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIE-FTKKPNCVESSKWLI 123
+ N C VVQ EV NG PV F+ G+I T T ++I+ F K C S W
Sbjct: 24 TGNKSCPPNVVQAKLEVWNGTPVTFTPYNAKDGVILTSTDLNIKSFPIKSPCGPSFVW-- 197
Query: 124 FVDSVIQKACVGI-----GGPENYPGFRTLSGTFNIEKHESGFGYRLGYC---VKDSPTC 175
+QK G+ GG E PG T+ F IEK +G Y L +C K + C
Sbjct: 198 ----KLQKELTGVWFLVTGGVEGNPGADTIVNWFKIEK--AGKDYVLSFCPSVCKCNTLC 359
Query: 176 LDIG 179
++G
Sbjct: 360 RELG 371
>TC15509 weakly similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor,
partial (77%)
Length = 797
Score = 49.3 bits (116), Expect = 5e-07
Identities = 41/120 (34%), Positives = 53/120 (44%), Gaps = 3/120 (2%)
Frame = +1
Query: 7 LTLSFL-LFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLGKS 65
L L FL L V T LA ++ +D+ GNPI PG KYY+ P GGL LG +
Sbjct: 25 LPLCFLSLLVLNTQPLLA--DEPEPAAVDMQGNPILPGVKYYVRPV--SAENGGLALGHT 192
Query: 66 SNSDCEVTVVQDYNEVINGVPVKFSIPEISP-GIIFTGTPIDIEF-TKKPNCVESSKWLI 123
N C + ++ D + G P F S G I T + +E K C E W I
Sbjct: 193 RNKTCPLDIILDPFSL--GTPTVFQTSSSSDLGYIPISTDLTVEMPVLKSPCKEPKVWRI 366
>TC15508 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(66%)
Length = 877
Score = 48.1 bits (113), Expect = 1e-06
Identities = 36/117 (30%), Positives = 53/117 (44%), Gaps = 2/117 (1%)
Frame = +3
Query: 7 LTLSFL-LFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLGKS 65
L L FL L V T LA ++ V+D+ GNP+ PG +YY+ P GGL LG +
Sbjct: 48 LPLCFLSLLVLNTQPLLA--DEPEPAVVDMQGNPLLPGVQYYVRPV--SAEKGGLALGHT 215
Query: 66 SNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKKPN-CVESSKW 121
N C + ++ D + +I V + G I T + +E + C E W
Sbjct: 216 RNKTCPLDIILDPSSIIGSPTVFHTSSSSDVGYIPISTDLTVEMPILGSPCKEPKVW 386
>BP083453
Length = 382
Score = 26.6 bits (57), Expect = 3.6
Identities = 18/53 (33%), Positives = 24/53 (44%), Gaps = 9/53 (16%)
Frame = +3
Query: 114 NCVESSKW-----LIFVDSV----IQKACVGIGGPENYPGFRTLSGTFNIEKH 157
+C E KW +F+ I KA V IGG PG +S FN+ +H
Sbjct: 75 SCPELDKWNVEFNFLFISKSPSWDINKA-VSIGGQTMNPGLALMS*NFNLNRH 230
>AW720487
Length = 575
Score = 26.2 bits (56), Expect = 4.7
Identities = 15/47 (31%), Positives = 22/47 (45%)
Frame = -1
Query: 27 DAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLGKSSNSDCEVT 73
D ++ I GNPI PGG+ G + G L L K + ++T
Sbjct: 224 DRNSRLSPITGNPILPGGRL-------GAVNGDLCL*KREKRESDIT 105
>BP081457
Length = 390
Score = 25.8 bits (55), Expect = 6.2
Identities = 16/71 (22%), Positives = 29/71 (40%)
Frame = +2
Query: 96 PGIIFTGTPIDIEFTKKPNCVESSKWLIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIE 155
P + P+ + FTK P + ++F+ + + + R ++E
Sbjct: 128 PSLTPLDVPLPLNFTKYPRFQKKMTKILFIGKINEAGLFWALRRQ----LRAPERQLHVE 295
Query: 156 KHESGFGYRLG 166
KH SG +RLG
Sbjct: 296 KHASGPNWRLG 328
>TC16128 homologue to UP|PSE1_ARATH (Q9S831) Photosystem I reaction center
subunit IV A, chloroplast precursor (PSI-E A), partial
(57%)
Length = 605
Score = 25.8 bits (55), Expect = 6.2
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = -1
Query: 45 KYYILPAIRGPLGGGLRLGKSSNSDCEVTV 74
+ + L + GP+GGGL G +S S V V
Sbjct: 317 RIFTLEPLLGPIGGGLGFGAASPSAVGVAV 228
>TC7942 similar to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragment) ,
partial (91%)
Length = 1288
Score = 25.4 bits (54), Expect = 8.0
Identities = 23/90 (25%), Positives = 35/90 (38%), Gaps = 23/90 (25%)
Frame = +3
Query: 36 NGNPIFPGGK-----------------------YYILPAIRGPLGGGLRLGKSSNSDCEV 72
+G PIFP G+ YY+ + GGL ++ N + E+
Sbjct: 549 SGPPIFPKGEYGTGFNPNTPETLPSWLSQQDLDYYVSKFNKTGFSGGLNYYRNLNLNWEL 728
Query: 73 TVVQDYNEVINGVPVKFSIPEISPGIIFTG 102
T + N VPVKF + ++ FTG
Sbjct: 729 TAPWTGVGITN-VPVKFILGDVDITYNFTG 815
>TC14559 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated
protein-like, partial (92%)
Length = 1125
Score = 25.4 bits (54), Expect = 8.0
Identities = 14/39 (35%), Positives = 18/39 (45%)
Frame = +1
Query: 4 VLSLTLSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFP 42
+LSL L L +FT + N V+Q D N P P
Sbjct: 52 LLSLLLFIL*SIFTFTIDFHLQNGEVQQQRDRNPEPTHP 168
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,772,301
Number of Sequences: 28460
Number of extensions: 54332
Number of successful extensions: 233
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 223
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 223
length of query: 220
length of database: 4,897,600
effective HSP length: 87
effective length of query: 133
effective length of database: 2,421,580
effective search space: 322070140
effective search space used: 322070140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC122730.10


