

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122729.4 - phase: 0
(113 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
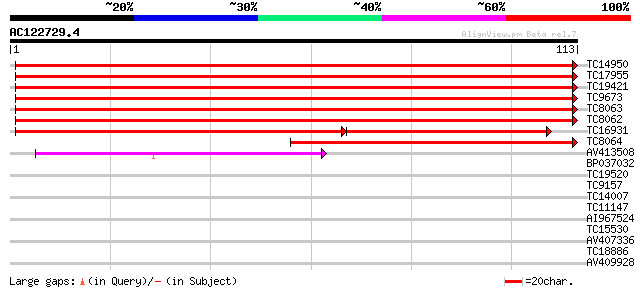
Score E
Sequences producing significant alignments: (bits) Value
TC14950 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {M... 185 8e-49
TC17955 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {M... 185 8e-49
TC19421 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {M... 183 4e-48
TC9673 UP|BAA31218 (BAA31218) Histone H3, complete 178 1e-46
TC8063 UP|BAA31218 (BAA31218) Histone H3, complete 178 1e-46
TC8062 UP|BAA31218 (BAA31218) Histone H3, complete 178 1e-46
TC16931 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {M... 116 7e-42
TC8064 homologue to GB|AAB36496.1|488573|MSU09463 histone H3.2 p... 84 3e-18
AV413508 79 8e-17
BP037032 27 0.49
TC19520 similar to UP|O22806 (O22806) Expressed protein, partial... 25 2.4
TC9157 25 2.4
TC14007 similar to UP|Q9FI93 (Q9FI93) Genomic DNA, chromosome 5,... 24 3.2
TC11147 similar to UP|Q9ZSQ7 (Q9ZSQ7) Protein phosphatase 2C hom... 24 3.2
AI967524 23 5.4
TC15530 similar to UP|AAQ84318 (AAQ84318) Fiber protein Fb27, pa... 23 5.4
AV407336 23 7.0
TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/h... 23 7.0
AV409928 23 7.0
>TC14950 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {Medicago
sativa;}, complete
Length = 641
Score = 185 bits (470), Expect = 8e-49
Identities = 97/112 (86%), Positives = 102/112 (90%)
Frame = +2
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 137 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 316
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAVSALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 317 FQSSAVSALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 472
>TC17955 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {Medicago
sativa;}, complete
Length = 639
Score = 185 bits (470), Expect = 8e-49
Identities = 97/112 (86%), Positives = 102/112 (90%)
Frame = +1
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 130 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 309
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAVSALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 310 FQSSAVSALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 465
>TC19421 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {Medicago
sativa;}, complete
Length = 672
Score = 183 bits (464), Expect = 4e-48
Identities = 96/112 (85%), Positives = 101/112 (89%)
Frame = +3
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 165 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 344
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAVSALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA IR ERA
Sbjct: 345 FQSSAVSALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARXIRGERA 500
>TC9673 UP|BAA31218 (BAA31218) Histone H3, complete
Length = 690
Score = 178 bits (452), Expect = 1e-46
Identities = 93/112 (83%), Positives = 99/112 (88%)
Frame = +1
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 148 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 327
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 328 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 483
>TC8063 UP|BAA31218 (BAA31218) Histone H3, complete
Length = 723
Score = 178 bits (452), Expect = 1e-46
Identities = 93/112 (83%), Positives = 99/112 (88%)
Frame = +2
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 143 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 322
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 323 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 478
>TC8062 UP|BAA31218 (BAA31218) Histone H3, complete
Length = 768
Score = 178 bits (452), Expect = 1e-46
Identities = 93/112 (83%), Positives = 99/112 (88%)
Frame = +3
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 123 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 302
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 303 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 458
>TC16931 PIR|S04520|S04520 histone H3 (clone pH3c-1) - alfalfa {Medicago
sativa;}, partial (96%)
Length = 440
Score = 116 bits (290), Expect(2) = 7e-42
Identities = 59/66 (89%), Positives = 61/66 (92%)
Frame = +1
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 118 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 297
Query: 62 FQISAV 67
FQ SAV
Sbjct: 298 FQSSAV 315
Score = 67.8 bits (164), Expect(2) = 7e-42
Identities = 34/41 (82%), Positives = 37/41 (89%)
Frame = +2
Query: 68 SALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRI 108
SALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RI
Sbjct: 317 SALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRI 439
>TC8064 homologue to GB|AAB36496.1|488573|MSU09463 histone H3.2 precursor
{Medicago sativa;} , partial (45%)
Length = 486
Score = 84.0 bits (206), Expect = 3e-18
Identities = 44/57 (77%), Positives = 49/57 (85%)
Frame = +1
Query: 57 KTDLRFQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
+TDLRFQ AV ALQE AEA +VGLFED +LCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 79 QTDLRFQSHAVLALQEAAEAYLVGLFEDTHLCAIHAKRVTIMPKDIQLARRIRGERA 249
>AV413508
Length = 265
Score = 79.3 bits (194), Expect = 8e-17
Identities = 47/86 (54%), Positives = 51/86 (58%), Gaps = 28/86 (32%)
Frame = +3
Query: 6 SALATGGMKKPHRFRPGTVALR----------------------------EIRKYQKITE 37
SA TGG+KKPHR+RPGTVALR EIRKYQK TE
Sbjct: 3 SAPTTGGVKKPHRYRPGTVALR*FTVFPQILSRFLSVAED*IFLFVYNFSEIRKYQKSTE 182
Query: 38 LLIRKLPFKRLVHEIAQDFKTDLRFQ 63
LLIRKLPF+RLV EIAQDFK L F+
Sbjct: 183LLIRKLPFQRLVREIAQDFKVILVFK 260
>BP037032
Length = 577
Score = 26.9 bits (58), Expect = 0.49
Identities = 12/36 (33%), Positives = 19/36 (52%)
Frame = +1
Query: 16 PHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHE 51
PHR RP V + + + + LIR LP R++ +
Sbjct: 193 PHRLRPRNVRVHTVSDQKPLGSELIRILPHVRILRQ 300
>TC19520 similar to UP|O22806 (O22806) Expressed protein, partial (12%)
Length = 560
Score = 24.6 bits (52), Expect = 2.4
Identities = 19/83 (22%), Positives = 42/83 (49%), Gaps = 11/83 (13%)
Frame = -2
Query: 14 KKPHRFRPGTVALREIRKYQKITELLIRK--------LPFKRLVHEIAQDFKTDLR---F 62
K ++++P + +R +K+T L++K + F R ++ + + F LR F
Sbjct: 523 KFQYKYKP-KINVRVKLSREKLTRKLLKKRYHTAAMSINFLRFLNYLGRLFFGKLRNGFF 347
Query: 63 QISAVSALQEIAEACIVGLFEDM 85
S+VS + ++ +C++ ED+
Sbjct: 346 LSSSVSLFKSLSSSCLIFKLEDL 278
>TC9157
Length = 621
Score = 24.6 bits (52), Expect = 2.4
Identities = 9/30 (30%), Positives = 19/30 (63%)
Frame = -2
Query: 82 FEDMNLCAIHAKRVTIVPKDIKLACRIRCE 111
+++++LC H + VPK K++ R+ C+
Sbjct: 284 WDEIHLCPNHGG*ASEVPKGTKISPRVTCK 195
>TC14007 similar to UP|Q9FI93 (Q9FI93) Genomic DNA, chromosome 5, TAC
clone:K18J17, partial (16%)
Length = 570
Score = 24.3 bits (51), Expect = 3.2
Identities = 10/32 (31%), Positives = 20/32 (62%)
Frame = -3
Query: 24 VALREIRKYQKITELLIRKLPFKRLVHEIAQD 55
++ +++ +YQ++ ELL+R L V +QD
Sbjct: 139 ISTKDLHQYQRLYELLLRTLKRSSFVS*SSQD 44
>TC11147 similar to UP|Q9ZSQ7 (Q9ZSQ7) Protein phosphatase 2C homolog,
partial (7%)
Length = 583
Score = 24.3 bits (51), Expect = 3.2
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = +1
Query: 78 IVGLFEDMNLCAIHAKRVTIVPK 100
IVGLF C +H KRV+I K
Sbjct: 91 IVGLFMVFVCC*VHNKRVSICSK 159
>AI967524
Length = 460
Score = 23.5 bits (49), Expect = 5.4
Identities = 21/61 (34%), Positives = 27/61 (43%), Gaps = 4/61 (6%)
Frame = -1
Query: 19 FRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLRFQI----SAVSALQEIA 74
FRP A RK + L R LPF V + D+K FQ+ + V A +IA
Sbjct: 196 FRPLNWAFISFRKRDQAGRRLFRTLPFS--VFIVFSDWKMLNCFQVVKAWNEVRASSDIA 23
Query: 75 E 75
E
Sbjct: 22 E 20
>TC15530 similar to UP|AAQ84318 (AAQ84318) Fiber protein Fb27, partial (59%)
Length = 692
Score = 23.5 bits (49), Expect = 5.4
Identities = 13/41 (31%), Positives = 20/41 (48%)
Frame = -3
Query: 4 RKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLP 44
++ ALA G KP F + L E +KY+ E + +P
Sbjct: 324 KRPALAPLGGAKPKNFCKLVLILMEHKKYRNAMEQSVGSVP 202
>AV407336
Length = 418
Score = 23.1 bits (48), Expect = 7.0
Identities = 8/18 (44%), Positives = 12/18 (66%)
Frame = -3
Query: 73 IAEACIVGLFEDMNLCAI 90
I +AC+ LF+D+ C I
Sbjct: 56 ICDACLFSLFQDVIFCLI 3
>TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (23%)
Length = 547
Score = 23.1 bits (48), Expect = 7.0
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = -2
Query: 95 VTIVPKDIKLACRIRCER 112
+T VPK + ++CRI ER
Sbjct: 429 LTFVPKLVPVSCRIPAER 376
>AV409928
Length = 414
Score = 23.1 bits (48), Expect = 7.0
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +2
Query: 26 LREIRKYQKITELLIRKLPFKRLVHEIAQD 55
LR + + I ELL +K+ +++VH I Q+
Sbjct: 101 LRTLGNIRLIGELLKQKMVPEKIVHHIVQE 190
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.328 0.140 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,608,425
Number of Sequences: 28460
Number of extensions: 16225
Number of successful extensions: 86
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 86
length of query: 113
length of database: 4,897,600
effective HSP length: 89
effective length of query: 24
effective length of database: 2,364,660
effective search space: 56751840
effective search space used: 56751840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 47 (22.7 bits)
Medicago: description of AC122729.4


