

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122727.11 + phase: 0 /pseudo/partial
(359 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
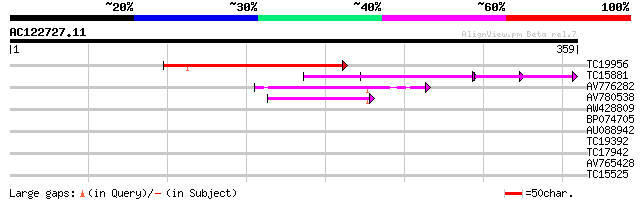
Score E
Sequences producing significant alignments: (bits) Value
TC19956 145 1e-35
TC15881 similar to GB|AAN18117.1|23308295|BT000548 At4g38160/F20... 53 7e-08
AV776282 45 2e-05
AV780538 44 4e-05
AW428809 28 1.8
BP074705 27 4.1
AU088942 27 5.3
TC19392 weakly similar to UP|Q9LR29 (Q9LR29) F26F24.17, partial ... 27 5.3
TC17942 weakly similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20... 27 6.9
AV765428 27 6.9
TC15525 UP|Q40209 (Q40209) RAB5A, complete 26 9.0
>TC19956
Length = 568
Score = 145 bits (365), Expect = 1e-35
Identities = 67/121 (55%), Positives = 96/121 (78%), Gaps = 4/121 (3%)
Frame = +2
Query: 98 ASSLYAGYARPSLS----EMKKDKATLRKVVYEFLRGIGIVPDELDGLELPVTVDVMKER 153
+SS + Y PS++ + +K+K R +++++L+G+GI+PDEL LELP TVDVM+ER
Sbjct: 206 SSSNFPEYEMPSVTWGVIQGRKEKLVSRVIIFDYLKGLGIIPDELHDLELPSTVDVMRER 385
Query: 154 VDFLHSLGLTIEDINNYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHAS 213
V+FL LGLT++DINNYPL+LGCSV+KNM+PVL YL K G+ +S + +F+++YPQVLHAS
Sbjct: 386 VEFLQKLGLTVDDINNYPLMLGCSVRKNMIPVLGYLEKXGISRSKLGEFVKSYPQVLHAS 565
Query: 214 V 214
V
Sbjct: 566 V 568
Score = 38.9 bits (89), Expect = 0.001
Identities = 30/104 (28%), Positives = 46/104 (43%), Gaps = 4/104 (3%)
Frame = +2
Query: 183 VPVLDYLGKLGVRKSTITQFLRTYPQVLHASVVVDLVPV----VKYLQGMDIKPDDIPRV 238
V + DYL LG+ P LH + V V V++LQ + + DDI
Sbjct: 290 VIIFDYLKGLGI-----------IPDELHDLELPSTVDVMRERVEFLQKLGLTVDDI--- 427
Query: 239 LERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILTRFPEIL 282
YP +LG + M + YL G+ R +LG + +P++L
Sbjct: 428 -NNYPLMLGCSVRKNMIPVLGYLEKXGISRSKLGEFVKSYPQVL 556
>TC15881 similar to GB|AAN18117.1|23308295|BT000548 At4g38160/F20D10_280
{Arabidopsis thaliana;}, partial (31%)
Length = 548
Score = 53.1 bits (126), Expect = 7e-08
Identities = 24/104 (23%), Positives = 53/104 (50%)
Frame = +3
Query: 223 KYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTSVAYLIGIGVGRRELGGILTRFPEIL 282
+YL+ + I+ +P ++ + P++L L + V L +G E+ + +FP IL
Sbjct: 219 EYLRSIGIQERKLPSIVSKCPKILALGLNEKIVPMVECLKTLGTKPHEVASAIAKFPHIL 398
Query: 283 GMRVGRVIKPFVEYLESLGIPRLAIARLIETQPYILGFDLDEKV 326
V + P + + ++LG+P I ++I P ++ + ++ K+
Sbjct: 399 SHSVEEKLCPLLAFFQALGVPEKQIGKMILLNPRLISYSIETKM 530
Score = 50.1 bits (118), Expect = 6e-07
Identities = 20/110 (18%), Positives = 57/110 (51%)
Frame = +3
Query: 187 DYLGKLGVRKSTITQFLRTYPQVLHASVVVDLVPVVKYLQGMDIKPDDIPRVLERYPEVL 246
+YL +G+++ + + P++L + +VP+V+ L+ + KP ++ + ++P +L
Sbjct: 219 EYLRSIGIQERKLPSIVSKCPKILALGLNEKIVPMVECLKTLGTKPHEVASAIAKFPHIL 398
Query: 247 GFKLEGTMSTSVAYLIGIGVGRRELGGILTRFPEILGMRVGRVIKPFVEY 296
+E + +A+ +GV +++G ++ P ++ + + V++
Sbjct: 399 SHSVEEKLCPLLAFFQALGVPEKQIGKMILLNPRLISYSIETKMAEIVDF 548
Score = 45.1 bits (105), Expect = 2e-05
Identities = 22/65 (33%), Positives = 36/65 (54%)
Frame = +3
Query: 295 EYLESLGIPRLAIARLIETQPYILGFDLDEKVKPNVKSLEEFNVRETSLASIIAQYPDII 354
EYL S+GI + ++ P IL L+EK+ P V+ L+ + +AS IA++P I+
Sbjct: 219 EYLRSIGIQERKLPSIVSKCPKILALGLNEKIVPMVECLKTLGTKPHEVASAIAKFPHIL 398
Query: 355 GTDLE 359
+E
Sbjct: 399 SHSVE 413
>AV776282
Length = 564
Score = 44.7 bits (104), Expect = 2e-05
Identities = 34/115 (29%), Positives = 58/115 (49%), Gaps = 4/115 (3%)
Frame = -2
Query: 156 FLHSLGLTIEDINNYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVV 215
FL S+G IE+++ + +L CSV+ +P LDY +G + T R +PQ S+
Sbjct: 506 FLPSIG--IEEVSKHTDLLSCSVEDKFLPRLDYFENIGFSRDDATSMFRRFPQSFCCSIK 333
Query: 216 VDLVPVVKYL---QGMDIKPDD-IPRVLERYPEVLGFKLEGTMSTSVAYLIGIGV 266
+L P +Y G ++K + I VL L K+E +S + +++ +GV
Sbjct: 332 NNLEPNYQYFVVEMGREMKETERISTVLSH----LA*KIE--LSLDINHVLELGV 186
>AV780538
Length = 503
Score = 43.9 bits (102), Expect = 4e-05
Identities = 23/71 (32%), Positives = 35/71 (48%), Gaps = 3/71 (4%)
Frame = -1
Query: 164 IEDINNYPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVVVDLVPVVK 223
IE+++ +P +L CSV+ +P LDY +G T R +PQ+ S+ L P
Sbjct: 500 IEEVSKHPDLLSCSVEDKFLPRLDYFENIGFSLDDATSMFRRFPQLFCCSIKNKLEPQSH 321
Query: 224 YL---QGMDIK 231
Y G D+K
Sbjct: 320 YFVVEMGRDLK 288
>AW428809
Length = 476
Score = 28.5 bits (62), Expect = 1.8
Identities = 18/55 (32%), Positives = 27/55 (48%), Gaps = 4/55 (7%)
Frame = +1
Query: 170 YPLVLGCSVKKNMVPVLDYLGKLGVRKSTITQFLRTYPQVLHASVV----VDLVP 220
Y +V G ++ M P+L + LG+R + I+ L Y V+ VDLVP
Sbjct: 94 YVIVSGYALNVAMDPILIFRCNLGIRGAAISHVLSQYVMAFALFVILMKKVDLVP 258
>BP074705
Length = 379
Score = 27.3 bits (59), Expect = 4.1
Identities = 13/28 (46%), Positives = 19/28 (67%)
Frame = +1
Query: 178 VKKNMVPVLDYLGKLGVRKSTITQFLRT 205
VKKN+V +LD+LG + K+ FLR+
Sbjct: 4 VKKNLVLMLDFLGLF*LGKNKFGHFLRS 87
>AU088942
Length = 570
Score = 26.9 bits (58), Expect = 5.3
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Frame = +3
Query: 301 GIPRLAIARLIETQPYILGFDLDEKVKPNVKSLEEFNVRETSL--ASIIAQYPDIIGTDL 358
G+PR + A +I+ PYI D VK V L +R +S A + +P + ++L
Sbjct: 105 GLPRSSDAEIIDALPYIDDDYGDPXVKVEVDRLVXDEMRRSSKKPADFLKDFPPLPTSNL 284
Query: 359 E 359
E
Sbjct: 285 E 287
>TC19392 weakly similar to UP|Q9LR29 (Q9LR29) F26F24.17, partial (24%)
Length = 491
Score = 26.9 bits (58), Expect = 5.3
Identities = 15/56 (26%), Positives = 28/56 (49%), Gaps = 4/56 (7%)
Frame = +2
Query: 168 NNYPLVLGCSVKKNMVPVL----DYLGKLGVRKSTITQFLRTYPQVLHASVVVDLV 219
N+Y L+LG + NMV L ++ + + ITQ + +P + H ++ L+
Sbjct: 218 NDYVLILGDGISNNMVKPLNGTFEFWQHVPTHVAGITQVTQKFPPLNHHRALITLL 385
>TC17942 weakly similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20, partial
(8%)
Length = 523
Score = 26.6 bits (57), Expect = 6.9
Identities = 12/34 (35%), Positives = 20/34 (58%)
Frame = +3
Query: 51 CNYIRKIRFITKLQCSVAGRTDSYRTSAADSQGS 84
C + R F+ K +CSV GRT S+ + D++ +
Sbjct: 138 CVFTRNFCFLVKNKCSVYGRTLSHISVEFDNEAA 239
>AV765428
Length = 422
Score = 26.6 bits (57), Expect = 6.9
Identities = 16/41 (39%), Positives = 19/41 (46%)
Frame = -2
Query: 265 GVGRRELGGILTRFPEILGMRVGRVIKPFVEYLESLGIPRL 305
GVGR G T F + G R G + +PF Y IP L
Sbjct: 415 GVGRTLKGRT*TGFGMLCGARPGSMDEPFDPYAFQFNIPLL 293
>TC15525 UP|Q40209 (Q40209) RAB5A, complete
Length = 1176
Score = 26.2 bits (56), Expect = 9.0
Identities = 13/36 (36%), Positives = 21/36 (58%)
Frame = -2
Query: 222 VKYLQGMDIKPDDIPRVLERYPEVLGFKLEGTMSTS 257
+KY +KP + RV ++ P +LGF+L + TS
Sbjct: 866 LKYFMNNMMKP*LLRRVYQQEPSLLGFELAEPLVTS 759
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.141 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,192,182
Number of Sequences: 28460
Number of extensions: 63798
Number of successful extensions: 298
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 298
length of query: 359
length of database: 4,897,600
effective HSP length: 91
effective length of query: 268
effective length of database: 2,307,740
effective search space: 618474320
effective search space used: 618474320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC122727.11


