

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122725.8 - phase: 0 /pseudo
(297 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
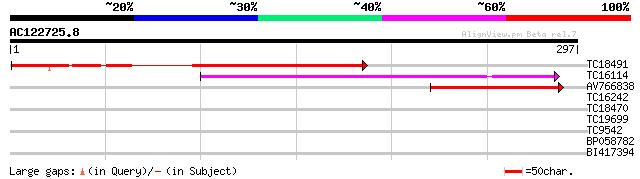
TC18491 similar to UP|RRF_DAUCA (P37706) Ribosome recycling fact... 168 1e-42
TC16114 similar to UP|AAQ62433 (AAQ62433) At3g01800, partial (73%) 123 3e-29
AV766838 101 1e-22
TC16242 UP|Q8W538 (Q8W538) Ribosomal S15 protein (Fragment), com... 35 0.020
TC18470 similar to PIR|T05606|T05606 protein kinase homolog F9D1... 28 1.9
TC19699 27 3.2
TC9542 homologue to UP|CAMT_MEDSA (Q40313) Caffeoyl-CoA O-methyl... 27 4.1
BP058782 26 7.1
BI417394 26 9.2
>TC18491 similar to UP|RRF_DAUCA (P37706) Ribosome recycling factor,
chloroplast precursor (Ribosome releasing factor,
chloroplast) (Nuclear located protein D2) (Fragment),
partial (43%)
Length = 474
Score = 168 bits (425), Expect = 1e-42
Identities = 102/190 (53%), Positives = 124/190 (64%), Gaps = 4/190 (2%)
Frame = +3
Query: 2 ATSFSSTTPLRSIFQNLP----IVPKALLSAHSSFTIKRSSTYVTLKLSGSGRHSLSKGL 57
ATSFS TP+RSIFQN P +P++ S TI S++YVTLKLS + +LS
Sbjct: 6 ATSFSPATPVRSIFQNPPKTTLFLPESFYGVKRS-TISASASYVTLKLSRT--LTLSAKP 176
Query: 58 LTLHTRYQNYYSRSACFQF*TTSLVFIVICFF*KIPRKGFVRAATIEEIEAEKASIENDV 117
+ H R G VRAAT+EEIEAEK +IE DV
Sbjct: 177 ILSHRRM-------------------------------GIVRAATMEEIEAEKKAIEKDV 263
Query: 118 KSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLVSP 177
++RME+T++NV++NF+SIRTGR+ PSMLDKIEVEYYG+P SLK+IAQISTPD SSLLV P
Sbjct: 264 RNRMEKTIENVRSNFNSIRTGRSNPSMLDKIEVEYYGAPASLKSIAQISTPDGSSLLVQP 443
Query: 178 YDKSSLKAIE 187
YDKSSLKAIE
Sbjct: 444 YDKSSLKAIE 473
>TC16114 similar to UP|AAQ62433 (AAQ62433) At3g01800, partial (73%)
Length = 1297
Score = 123 bits (309), Expect = 3e-29
Identities = 70/188 (37%), Positives = 110/188 (58%)
Frame = +3
Query: 101 ATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLK 160
+TIE +I+ + S+ME + + T S +RTGRA+P MLD I VE G + L
Sbjct: 357 STIEVPLVLGPTIKANAVSQMEAAISALSTELSKLRTGRASPGMLDHIIVETGGVKIPLV 536
Query: 161 TIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKE 220
IA +S D +L V+PYD +LK +E AIV+S +G+ P DGE + +IP LT + +
Sbjct: 537 RIALVSVIDPKTLSVNPYDPQTLKQLENAIVSSPLGLNPKADGERLIAAIPPLTKEHMQA 716
Query: 221 LTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVD 280
+ K+V+K E+ + ++R R+ A+ A +KL + L +D++K L ++ LT +IK +
Sbjct: 717 MNKLVTKSCEDARQSIRRARQKAMDAIKKL--NSSLPKDDIKRLEKEVDDLTKKFIKTAE 890
Query: 281 ALYKQKEK 288
K KEK
Sbjct: 891 DTCKAKEK 914
>AV766838
Length = 493
Score = 101 bits (252), Expect = 1e-22
Identities = 50/70 (71%), Positives = 62/70 (88%)
Frame = -3
Query: 221 LTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVD 280
L+K+V ++GKVALRNI++DALKAYEKLEK+KKLSEDNVKDLSSDLQKL D+YIKKV+
Sbjct: 491 LSKVVFNPTKKGKVALRNIKKDALKAYEKLEKEKKLSEDNVKDLSSDLQKLPDEYIKKVE 312
Query: 281 ALYKQKEKVL 290
+ +QK+K L
Sbjct: 311 PISQQKKKEL 282
>TC16242 UP|Q8W538 (Q8W538) Ribosomal S15 protein (Fragment), complete
Length = 581
Score = 34.7 bits (78), Expect = 0.020
Identities = 21/65 (32%), Positives = 38/65 (58%), Gaps = 2/65 (3%)
Frame = -2
Query: 115 NDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSP--VSLKTIAQISTPDASS 172
N +++R E++L + + S R R+TP L+ + V ++G+P S+ T A +T + S
Sbjct: 193 NLLRARAEKSLTSSSVDISK-RASRSTPRKLNFLNVLFFGTPAATSVSTSAMAATLNTRS 17
Query: 173 LLVSP 177
LL+ P
Sbjct: 16 LLLLP 2
>TC18470 similar to PIR|T05606|T05606 protein kinase homolog F9D16.210 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(9%)
Length = 559
Score = 28.1 bits (61), Expect = 1.9
Identities = 10/21 (47%), Positives = 15/21 (70%)
Frame = +2
Query: 73 CFQF*TTSLVFIVICFF*KIP 93
CF+F T S++ I+ CF K+P
Sbjct: 392 CFKFCTRSMIAIIFCFIFKLP 454
>TC19699
Length = 511
Score = 27.3 bits (59), Expect = 3.2
Identities = 11/26 (42%), Positives = 18/26 (68%)
Frame = -2
Query: 126 DNVKTNFSSIRTGRATPSMLDKIEVE 151
DN+K NFS+ RT R + L+K +++
Sbjct: 312 DNIK*NFSTYRTSRLQENNLEKTKID 235
>TC9542 homologue to UP|CAMT_MEDSA (Q40313) Caffeoyl-CoA
O-methyltransferase (Trans-caffeoyl-CoA
3-O-methyltransferase) (CCoAMT) (CCoAOMT) , complete
Length = 1012
Score = 26.9 bits (58), Expect = 4.1
Identities = 11/37 (29%), Positives = 21/37 (56%)
Frame = -2
Query: 5 FSSTTPLRSIFQNLPIVPKALLSAHSSFTIKRSSTYV 41
F + LRS+F+N P + ++ H F +K S+++
Sbjct: 327 FLALISLRSMFKNCPSSAEVVMMFHGCFAVKSLSSFM 217
>BP058782
Length = 363
Score = 26.2 bits (56), Expect = 7.1
Identities = 14/59 (23%), Positives = 29/59 (48%)
Frame = +3
Query: 139 RATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGM 197
+A PS+L ++ + + T+ +TP+ SS + SS +++ + + VGM
Sbjct: 141 QALPSVLGSWQLLVFSKSILSSTLNSSATPNLSSSFLDSRAASSAMSVKLVLYSFKVGM 317
>BI417394
Length = 598
Score = 25.8 bits (55), Expect = 9.2
Identities = 11/32 (34%), Positives = 21/32 (65%)
Frame = +3
Query: 5 FSSTTPLRSIFQNLPIVPKALLSAHSSFTIKR 36
FSS+TP ++I++ I+P + +S S++ R
Sbjct: 138 FSSSTPSKNIYRKRIILPVSKISIQESYSNSR 233
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.133 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,827,505
Number of Sequences: 28460
Number of extensions: 45007
Number of successful extensions: 225
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 222
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 224
length of query: 297
length of database: 4,897,600
effective HSP length: 90
effective length of query: 207
effective length of database: 2,336,200
effective search space: 483593400
effective search space used: 483593400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC122725.8


