

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.4 + phase: 0
(307 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
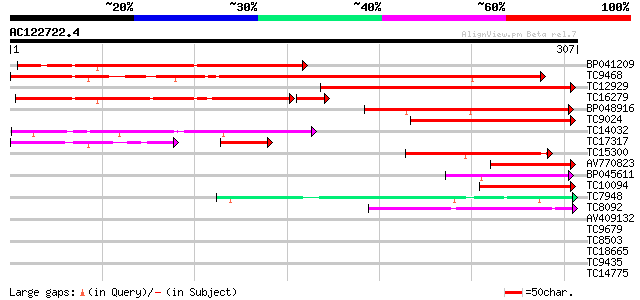
Score E
Sequences producing significant alignments: (bits) Value
BP041209 204 2e-53
TC9468 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (72%) 200 3e-52
TC12929 weakly similar to UP|Q9LV43 (Q9LV43) Genomic DNA, chromo... 165 1e-41
TC16279 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) correspon... 143 2e-39
BP048916 134 1e-32
TC9024 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) correspond... 97 3e-21
TC14032 homologue to UP|P93747 (P93747) At2g41990 protein, parti... 80 6e-16
TC17317 homologue to UP|Q84VY4 (Q84VY4) At5g42860, partial (20%) 58 7e-15
TC15300 weakly similar to UP|P93747 (P93747) At2g41990 protein, ... 57 4e-09
AV770823 55 2e-08
BP045611 49 1e-06
TC10094 similar to UP|Q84VY4 (Q84VY4) At5g42860, partial (10%) 45 1e-05
TC7948 43 8e-05
TC8092 similar to UP|Q8MLI6 (Q8MLI6) CG30127-PA, partial (3%) 40 5e-04
AV409132 38 0.002
TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, p... 35 0.012
TC8503 similar to GB|AAD30253.1|4835787|F25C20 ESTs gb|R65381 an... 33 0.061
TC18665 similar to UP|O23061 (O23061) BAC IG005I10, partial (65%) 30 0.51
TC9435 similar to UP|Q9AXJ7 (Q9AXJ7) Phosphatase-like protein Mt... 30 0.67
TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest c... 29 0.88
>BP041209
Length = 476
Score = 204 bits (518), Expect = 2e-53
Identities = 111/163 (68%), Positives = 126/163 (77%), Gaps = 6/163 (3%)
Frame = +1
Query: 5 TDSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP-----SPMDSPSHQSYGH 59
TDSDVTSF SSP + Y VQSPSR+S HDG+KS +M SPM+SPSH SYGH
Sbjct: 1 TDSDVTSFGASSP---PRPVYIVQSPSRDS-HDGEKSPSMHATPASNSPMESPSHPSYGH 168
Query: 60 HSRASSSSRISGGYN-SSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVSRRV 118
HSR SSSSR+SG YN SS LG+K +RK ND+GW KVIEEE GGYGDF + G+SRR
Sbjct: 169 HSRESSSSRVSGSYNNSSVLGKKGDRKGNDRGWPECKVIEEE-GGYGDFYREDKGLSRRT 345
Query: 119 QFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYF 161
QFF+A+VGFVLVF+LFC IIWAAS YKPQLSVKS+TVHNFYF
Sbjct: 346 QFFLAIVGFVLVFTLFCLIIWAASSPYKPQLSVKSVTVHNFYF 474
>TC9468 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (72%)
Length = 1317
Score = 200 bits (508), Expect = 3e-52
Identities = 124/302 (41%), Positives = 184/302 (60%), Gaps = 12/302 (3%)
Frame = +3
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTP-YYVQSPSRESSHDGDKS------STMQPSPMDSP 52
MH KTDS+VTS SSP RS + P Y+VQSPSR+S HDG+K+ ST SP SP
Sbjct: 48 MHAKTDSEVTSISASSPARSPPRRPLYFVQSPSRDS-HDGEKTVTTSFHSTPVLSPNASP 224
Query: 53 SHQSYGHHSRASSSSRISGGYNSSFLGRKVNRKVND--KGWIGSKVIEEENGGYGDFDGD 110
H +SSS+R S + +K N + K W VIEEE G+ D D
Sbjct: 225 PH--------SSSSTRFSAPHR-----KKDNHHHHHSLKPWKQIDVIEEEGLLQGE-DRD 362
Query: 111 GNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
+SRR F + ++GF+L+F+LF I+W AS KP++ +KS+ + GSD TGV
Sbjct: 363 RT-LSRRCYFLIFLLGFLLLFTLFSLILWGASRPMKPKIFIKSIKFDHVQVQAGSDSTGV 539
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSV 230
T M+++N +++ N TFFG++V+S + L YS++ +A G +K+++Q R+S R VSV
Sbjct: 540 ATDMISMNSTLKFTYRNTGTFFGVHVASTPLELSYSEIVIAAGNMKEFYQHRRSHRLVSV 719
Query: 231 NIQGSKVPLYGAGADVASS--VDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAV 288
+ G+K+PLYG+GA ++S+ + + L L F ++SR V+GKLV+ K+ +R+ CSI +
Sbjct: 720 AVMGNKIPLYGSGASLSSTTGMPTVPVPLNLNFVLRSRAYVLGKLVKPKYYKRIQCSITL 899
Query: 289 DP 290
DP
Sbjct: 900 DP 905
>TC12929 weakly similar to UP|Q9LV43 (Q9LV43) Genomic DNA, chromosome 3, P1
clone: MOB24, partial (18%)
Length = 609
Score = 165 bits (417), Expect = 1e-41
Identities = 80/139 (57%), Positives = 106/139 (75%), Gaps = 1/139 (0%)
Frame = +2
Query: 169 GVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTV 228
GVPTK+LTVN ++RM ++NPATFFGI+V S +NL++SD++VATGELKKY+Q RKS R V
Sbjct: 2 GVPTKILTVNSTLRMSIYNPATFFGIHVHSTPINLVFSDISVATGELKKYYQPRKSHRMV 181
Query: 229 SVNIQGSKVPLYGAGADVASSVDN-GKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIA 287
SVN+ G KVPLYGAG+ + S +SL L FE+ SRGNVVGKLVRTKH + ++C +
Sbjct: 182 SVNLVGKKVPLYGAGSTITESQTGVVVVSLKLKFEIVSRGNVVGKLVRTKHHKEITCPLV 361
Query: 288 VDPHNNKYIKLKENECRYN 306
+D +K I+ K+N C Y+
Sbjct: 362 IDSSGSKPIQFKKNSCTYD 418
>TC16279 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) corresponds to a
region of the predicted gene, partial (38%)
Length = 866
Score = 143 bits (361), Expect(2) = 2e-39
Identities = 89/158 (56%), Positives = 107/158 (67%), Gaps = 7/158 (4%)
Frame = +1
Query: 4 KTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKSSTMQP-----SPMDSPSHQSY 57
K+DSDVTS PSSP RS K+ YYVQSPSR+S HDGDKSS+MQ SPM+SPSH S+
Sbjct: 349 KSDSDVTSLAPSSPSRSPKRPVYYVQSPSRDS-HDGDKSSSMQATPISNSPMESPSHPSF 525
Query: 58 GHHSRASSSSRISGGYNSSFLGRKVNR-KVNDKGWIGSKVIEEENGGYGDFDGDGNGVSR 116
G HSR SS+SR SG + SS GRK R K NDKGW VI EE G Y +F + R
Sbjct: 526 GRHSRNSSASRFSGIFRSS-SGRKSGRGKRNDKGWPECDVILEE-GSYHEF--EDRAFMR 693
Query: 117 RVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSL 154
R Q +AV FV+VF++FC IIW AS +K +++VK L
Sbjct: 694 RCQGLIAVFTFVVVFTVFCLIIWGASRPFKAEVAVKKL 807
Score = 35.0 bits (79), Expect(2) = 2e-39
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = +3
Query: 156 VHNFYFGEGSDVTGVPTK 173
+HN Y GEGSD TG+PTK
Sbjct: 813 LHNLYIGEGSDFTGIPTK 866
>BP048916
Length = 555
Score = 134 bits (338), Expect = 1e-32
Identities = 73/120 (60%), Positives = 86/120 (70%), Gaps = 7/120 (5%)
Frame = -2
Query: 193 GIYVSSKSVNLMYSDLTVATG-----ELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVA 247
G Y + LM LT TG ++KKYHQ+RKS TV VN+QGSKVPLYGAG ++A
Sbjct: 521 GKYAEESFLLLMLIMLTTLTGKFMLLQVKKYHQRRKSTSTVYVNLQGSKVPLYGAGVNLA 342
Query: 248 S--SVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNNKYIKLKENECRY 305
S VD+ KI + LVF+VKSRGNVVGKLV +KHT+ VSCS+ VD HNN IKLKEN CRY
Sbjct: 341 SLVEVDDRKIPVRLVFDVKSRGNVVGKLVTSKHTRHVSCSVPVDSHNNHPIKLKENACRY 162
>TC9024 similar to UP|Q9XE35 (Q9XE35) ESTD24835(R2634) corresponds to a
region of the predicted gene, partial (8%)
Length = 588
Score = 97.1 bits (240), Expect = 3e-21
Identities = 48/90 (53%), Positives = 62/90 (68%), Gaps = 1/90 (1%)
Frame = +3
Query: 218 YHQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDNGKI-SLTLVFEVKSRGNVVGKLVRT 276
Y+Q RKS R VSVN+ G KVPLYGAG+ + S + SL L FE+ SRGNVVGKLVRT
Sbjct: 3 YYQPRKSHRMVSVNLVGKKVPLYGAGSTITESQTGVVVVSLKLKFEIVSRGNVVGKLVRT 182
Query: 277 KHTQRVSCSIAVDPHNNKYIKLKENECRYN 306
KH + ++C + +D +K IK K+N C Y+
Sbjct: 183 KHHKEITCPLVIDSSGSKPIKFKKNSCTYD 272
>TC14032 homologue to UP|P93747 (P93747) At2g41990 protein, partial (20%)
Length = 572
Score = 79.7 bits (195), Expect = 6e-16
Identities = 64/182 (35%), Positives = 91/182 (49%), Gaps = 17/182 (9%)
Frame = +2
Query: 2 HIKTDSDVTS-----FDPSSPRSQKQTP-YYVQSPSRESSHDGDKSSTMQPSPMDSPSHQ 55
H K+DS+VTS S PRS + P YYVQSPS +HD +K S SP+ SP+H
Sbjct: 47 HQKSDSEVTSNSMEQSSSSPPRSPPRRPLYYVQSPS---NHDVEKMS-YGSSPVGSPAHH 214
Query: 56 SYG-------HHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFD 108
+ HHSR SS+SR S + R N + W K + + D D
Sbjct: 215 HFQYYLSSPIHHSRESSTSRFSASLKNPLPLRHQNHQPLSS-W---KKLHHQTDDDLDDD 382
Query: 109 GDGNGV---SRRVQ-FFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
+ + V +R ++ +F + FVL+F+ F I+W AS YKP++ +KS+ N G
Sbjct: 383 DENDAVYDSTRNIRLYFCFFLLFVLLFTTFSLILWGASKSYKPRVIIKSIVFENLNVQSG 562
Query: 165 SD 166
SD
Sbjct: 563 SD 568
>TC17317 homologue to UP|Q84VY4 (Q84VY4) At5g42860, partial (20%)
Length = 566
Score = 58.2 bits (139), Expect(2) = 7e-15
Identities = 44/97 (45%), Positives = 52/97 (53%), Gaps = 6/97 (6%)
Frame = +2
Query: 1 MHIKTDSDVTSFDPSS-PRSQKQTPYYVQSPSRESSHDGDKS-----STMQPSPMDSPSH 54
MH KTDS+VTS D SS RS ++ YYVQSP SHDG+K+ ST SPM SP H
Sbjct: 137 MHAKTDSEVTSLDASSTTRSPRRPAYYVQSP----SHDGEKTTTSFHSTPVISPMGSPPH 304
Query: 55 QSYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGW 91
SSSSR SG S + +R + K W
Sbjct: 305 SH-------SSSSRFSG---SRKMNHHNHRNNSTKPW 385
Score = 38.1 bits (87), Expect(2) = 7e-15
Identities = 16/28 (57%), Positives = 20/28 (71%)
Frame = +3
Query: 115 SRRVQFFVAVVGFVLVFSLFCFIIWAAS 142
SRR FF ++GF L+FSLF I+W AS
Sbjct: 447 SRRFYFFAFILGFFLLFSLFALILWGAS 530
>TC15300 weakly similar to UP|P93747 (P93747) At2g41990 protein, partial
(23%)
Length = 530
Score = 57.0 bits (136), Expect = 4e-09
Identities = 28/83 (33%), Positives = 52/83 (61%), Gaps = 3/83 (3%)
Frame = +1
Query: 215 LKKYHQQRKSRRTVSVNIQGSKVPLYGAGAD---VASSVDNGKISLTLVFEVKSRGNVVG 271
+K+++ +R S R ++V + G +VPLYG ++ + +D + L L F V+SR ++G
Sbjct: 34 MKEFYLRRNSHRNLTVVVTGHQVPLYGGVSELGNIKEHLDRVALPLNLTFVVRSRAFILG 213
Query: 272 KLVRTKHTQRVSCSIAVDPHNNK 294
+LV++K +R+ CS+ + H NK
Sbjct: 214 RLVKSKFYRRIRCSVTL--HGNK 276
>AV770823
Length = 464
Score = 54.7 bits (130), Expect = 2e-08
Identities = 22/46 (47%), Positives = 34/46 (73%)
Frame = -2
Query: 261 FEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNNKYIKLKENECRYN 306
FE+ SRGNVVGKLVRTKH + ++C + +D ++ I+ ++N C Y+
Sbjct: 460 FEIVSRGNVVGKLVRTKHHKEITCPLVLDSSGSRPIQFQKNSCTYD 323
>BP045611
Length = 570
Score = 48.5 bits (114), Expect = 1e-06
Identities = 26/72 (36%), Positives = 40/72 (55%), Gaps = 3/72 (4%)
Frame = -1
Query: 237 VPLYGAGADVASSVDNGK---ISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNN 293
+PLYG GA ++S+ + L L V+SRG V+GKLV+ K +++ CS+ +DP
Sbjct: 567 IPLYGGGASLSSTSGGAPAEALPLKLRVMVRSRGYVLGKLVKPKFNKKIECSVVMDPKKM 388
Query: 294 KYIKLKENECRY 305
N+C Y
Sbjct: 387 GAPVSLVNKCIY 352
>TC10094 similar to UP|Q84VY4 (Q84VY4) At5g42860, partial (10%)
Length = 595
Score = 45.4 bits (106), Expect = 1e-05
Identities = 20/52 (38%), Positives = 32/52 (61%)
Frame = +2
Query: 255 ISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNNKYIKLKENECRYN 306
+ L L F ++SR V+GKLV+ K+ +R+ CSI +DP I + C+Y+
Sbjct: 29 VPLKLSFVIRSRAYVLGKLVKPKYYKRIECSITLDPQKINAIVSLKKSCKYD 184
>TC7948
Length = 1230
Score = 42.7 bits (99), Expect = 8e-05
Identities = 47/205 (22%), Positives = 78/205 (37%), Gaps = 10/205 (4%)
Frame = +2
Query: 113 GVSRRV--QFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
G RR + F +V F+ + + +IWA KP ++ +TV+ F
Sbjct: 374 GAKRRAFRRVFCCIVVFLFIVLVTILLIWAILRPTKPTFILQDVTVYAF--------NAT 529
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY-SDLTVATGELKKYHQQRKSRRTVS 229
LT N + + NP G+Y S + Y S + +Q K S
Sbjct: 530 VPNFLTTNFQVTLTSRNPNEHIGVYYDRLSTYVTYRSQQVTYRTAIPPSYQGHKEFDVWS 709
Query: 230 VNIQGSKVPL-----YGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSC 284
+ G+ VP+ G G D A NG + +T+ + + R VG V ++ V C
Sbjct: 710 PFVSGNSVPVAPFNFVGLGQDQA----NGNVFITVKVDGRVRWK-VGAFVSGRYHLYVRC 874
Query: 285 S--IAVDPHNNKYIKLKENECRYNI 307
I++ N + N +Y +
Sbjct: 875 PAYISLGSRGNGVFVGENNAVKYQV 949
>TC8092 similar to UP|Q8MLI6 (Q8MLI6) CG30127-PA, partial (3%)
Length = 1073
Score = 40.0 bits (92), Expect = 5e-04
Identities = 31/115 (26%), Positives = 54/115 (46%), Gaps = 2/115 (1%)
Frame = +3
Query: 195 YVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDN-G 253
Y+ SV + Y D + G L ++Q + ++G V L AG+D + V+
Sbjct: 528 YLKGSSVEMFYRDSRLCGGALPAFYQASNNVTVFKAVLKGDGVEL--AGSDRTALVNAVA 701
Query: 254 KISLTLVFEVKSRGNV-VGKLVRTKHTQRVSCSIAVDPHNNKYIKLKENECRYNI 307
K S+ L ++++ + VG + K T +V C +AVD K K+ +C Y +
Sbjct: 702 KRSVPLTMKLRAPVKIKVGSVKTWKITVKVDCDVAVDQLTAK-SKIVHKDCSYGL 863
>AV409132
Length = 405
Score = 38.1 bits (87), Expect = 0.002
Identities = 25/96 (26%), Positives = 41/96 (42%), Gaps = 3/96 (3%)
Frame = +1
Query: 110 DGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTG 169
+G + RR+ AVV F+ + L F+IW KP+ ++ TV F
Sbjct: 121 EGRQILRRI--LAAVVAFIFLILLIIFLIWIILRPTKPRFMLQDATVFAFNLSTQQHTPP 294
Query: 170 VPT---KMLTVNCSMRMLVHNPATFFGIYVSSKSVN 202
+ T LT+ + + HNP G+Y + V+
Sbjct: 295 LLTPTPNTLTLTLQVTLSSHNPNARIGVYYHALHVH 402
>TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial
(54%)
Length = 710
Score = 35.4 bits (80), Expect = 0.012
Identities = 41/172 (23%), Positives = 68/172 (38%), Gaps = 4/172 (2%)
Frame = +2
Query: 121 FVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCS 180
F + +++ + I W + V T+ F F TG T L + +
Sbjct: 188 FKLIFTALIIIGIAVLIFWLIVRPNVVKFHVTDATLTQFNF------TGNNT--LNYDLT 343
Query: 181 MRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSK-VPL 239
+ + V NP GIY MY D+ T +++Q K +S +G+ VPL
Sbjct: 344 LNITVRNPNKRLGIYYDYIEARAMYQDVRFDTEFFDRFYQGHKKTNVISRQFKGTHVVPL 523
Query: 240 YGAGADVASSVDNGKISLTLVFEVKSRGNVVGKL--VRTKHTQ-RVSCSIAV 288
GA+ ++ K +VK V KL +TK + +V+C + V
Sbjct: 524 ---GAEETKELNKDKEDGVYGIDVKMYLKVRFKLGVFKTKTLKPKVTCELRV 670
>TC8503 similar to GB|AAD30253.1|4835787|F25C20 ESTs gb|R65381 and
gb|T44635 come from this gene. {Arabidopsis thaliana;},
partial (26%)
Length = 1151
Score = 33.1 bits (74), Expect = 0.061
Identities = 28/76 (36%), Positives = 36/76 (46%), Gaps = 6/76 (7%)
Frame = -3
Query: 7 SDVTSFDP----SSPR--SQKQTPYYVQSPSRESSHDGDKSSTMQPSPMDSPSHQSYGHH 60
SD T +P SSP S ++TP V S SR H + P+P SP HQ G
Sbjct: 543 SDSTELNPEVSCSSPARSSARRTPRGVLSESRPDHHARGAGARSSPTP--SPDHQPAGPR 370
Query: 61 SRASSSSRISGGYNSS 76
+ A + RI GG + S
Sbjct: 369 TPA-TGHRIGGGCSCS 325
>TC18665 similar to UP|O23061 (O23061) BAC IG005I10, partial (65%)
Length = 451
Score = 30.0 bits (66), Expect = 0.51
Identities = 27/75 (36%), Positives = 35/75 (46%)
Frame = -1
Query: 10 TSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQPSPMDSPSHQSYGHHSRASSSSRI 69
+S DP P+SQ TPY P RES S T PSP SPS + + SS
Sbjct: 424 SSSDP--PQSQFPTPY----PRRESP-----SQTHPPSPSGSPSPPPSSCTDQTTQSSPS 278
Query: 70 SGGYNSSFLGRKVNR 84
S + ++ R+V R
Sbjct: 277 SHPPHKTWEERRVRR 233
>TC9435 similar to UP|Q9AXJ7 (Q9AXJ7) Phosphatase-like protein Mtc923,
partial (17%)
Length = 486
Score = 29.6 bits (65), Expect = 0.67
Identities = 24/74 (32%), Positives = 30/74 (40%)
Frame = +2
Query: 6 DSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQPSPMDSPSHQSYGHHSRASS 65
D D D SP S+ Q+P + SPS S PSP SPS Y SR S
Sbjct: 206 DDDENENDAVSP-SEPQSPDFYPSPSPSRS----------PSPSHSPSRAPYELRSRRSL 352
Query: 66 SSRISGGYNSSFLG 79
+ S + + G
Sbjct: 353 GKKPSSSSSGTRRG 394
>TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest chain
(isoform B1) - soybean (fragment) {Glycine
max;} , partial (22%)
Length = 848
Score = 29.3 bits (64), Expect = 0.88
Identities = 21/66 (31%), Positives = 30/66 (44%)
Frame = +1
Query: 11 SFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQPSPMDSPSHQSYGHHSRASSSSRIS 70
++ PSSPR +PY SP ++ S SP SPS +Y S +S +S
Sbjct: 148 AYSPSSPRLSPSSPYSPTSP----NYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSG--VS 309
Query: 71 GGYNSS 76
Y+ S
Sbjct: 310 PDYSPS 327
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,510,758
Number of Sequences: 28460
Number of extensions: 82691
Number of successful extensions: 799
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 765
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 781
length of query: 307
length of database: 4,897,600
effective HSP length: 90
effective length of query: 217
effective length of database: 2,336,200
effective search space: 506955400
effective search space used: 506955400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC122722.4


