

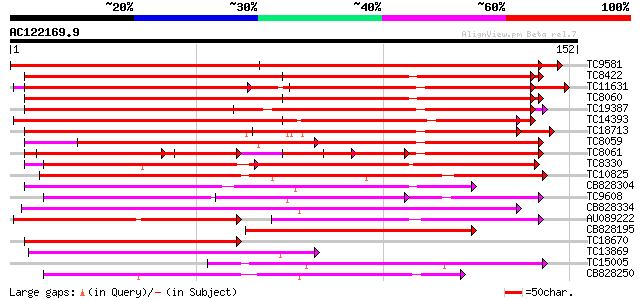
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.9 - phase: 0
(152 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete 237 7e-64
TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete 119 2e-28
TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-rela... 119 2e-28
TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regu... 119 3e-28
TC19387 weakly similar to UP|Q9FYK2 (Q9FYK2) F21J9.28, partial (... 115 3e-27
TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 106 2e-24
TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13... 105 4e-24
TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus ann... 103 2e-23
TC8061 UP|AAA34013 (AAA34013) Calmodulin, complete 52 1e-19
TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodu... 87 1e-18
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 83 2e-17
CB828304 68 8e-13
TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial ... 62 4e-11
CB828334 62 6e-11
AU089222 59 3e-10
CB828195 59 5e-10
TC18670 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 54 9e-09
TC13869 weakly similar to UP|Q9FR00 (Q9FR00) Avr9/Cf-9 rapidly e... 54 9e-09
TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partia... 53 2e-08
CB828250 49 3e-07
>TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete
Length = 1049
Score = 237 bits (604), Expect = 7e-64
Identities = 113/148 (76%), Positives = 133/148 (89%)
Frame = +3
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MD EL R+FQMFD+NGDGRITKKEL+DSL+NLGI I +++L QMIE+IDVNGDG VDID
Sbjct: 336 MDPTELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPDKELTQMIERIDVNGDGCVDID 515
Query: 61 EFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
EFGELYQ+IMDE+DEEEDM+EAFNVFDQNGDGFI+ EEL VL+SLG+K G+T+EDCK M
Sbjct: 516 EFGELYQSIMDERDEEEDMREAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKM 695
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAGAFAA 148
I KVDVDGDGMV++KEF+QMMK G F+A
Sbjct: 696 IMKVDVDGDGMVDYKEFKQMMKGGGFSA 779
Score = 50.8 bits (120), Expect = 1e-07
Identities = 26/76 (34%), Positives = 48/76 (62%)
Frame = +3
Query: 68 TIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVD 127
T + K + ++K F +FD+NGDG I+ +EL+ L +LG+ ++ MI+++DV+
Sbjct: 318 TSLIPKMDPTELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPD--KELTQMIERIDVN 491
Query: 128 GDGMVNFKEFQQMMKA 143
GDG V+ EF ++ ++
Sbjct: 492 GDGCVDIDEFGELYQS 539
>TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete
Length = 832
Score = 119 bits (299), Expect = 2e-28
Identities = 62/139 (44%), Positives = 91/139 (64%)
Frame = +1
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 169 EFKEAFSLFDKDGDGSITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 348
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++ +GFIS EL V+++LG K T E+ MI++
Sbjct: 349 LMARKMKDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 522
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG +N++EF ++M A
Sbjct: 523 DVDGDGQINYEEFVKVMMA 579
Score = 60.1 bits (144), Expect = 2e-10
Identities = 29/68 (42%), Positives = 46/68 (67%)
Frame = +1
Query: 74 DEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVN 133
D+ + KEAF++FD++GDG I+ +EL V+ SLG T + ++MI +VD DG+G ++
Sbjct: 157 DQISEFKEAFSLFDKDGDGSITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTID 330
Query: 134 FKEFQQMM 141
F EF +M
Sbjct: 331 FPEFLNLM 354
>TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-related protein
2, touch-induced, partial (68%)
Length = 530
Score = 119 bits (298), Expect = 2e-28
Identities = 62/137 (45%), Positives = 92/137 (66%)
Frame = +3
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E+ +IF FDKNGDG+I+ EL + L LG S E++ +M+ +ID NGDGY+D+ EF E
Sbjct: 99 EVRKIFNKFDKNGDGKISCTELKEMLCALGTKTSSEEVKRMMAEIDQNGDGYIDLKEFAE 278
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
+ E D+ +++++A +++D + +G IS EL +VL LG K +L DC+ MI V
Sbjct: 279 FHLHDAGE-DDSKELRDACDLYDLDKNGLISANELHSVLKKLGEK--CSLGDCRKMISNV 449
Query: 125 DVDGDGMVNFKEFQQMM 141
D DGDG VNF+EF++MM
Sbjct: 450 DADGDGNVNFEEFKKMM 500
Score = 55.8 bits (133), Expect = 3e-09
Identities = 29/75 (38%), Positives = 47/75 (62%)
Frame = +3
Query: 76 EEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVNFK 135
++++++ FN FD+NGDG IS EL +L +LG K + E+ K M+ ++D +GDG ++ K
Sbjct: 93 DDEVRKIFNKFDKNGDGKISCTELKEMLCALGTK--TSSEEVKRMMAEIDQNGDGYIDLK 266
Query: 136 EFQQMMKAGAFAADS 150
EF + A DS
Sbjct: 267 EFAEFHLHDAGEDDS 311
Score = 40.8 bits (94), Expect = 1e-04
Identities = 22/64 (34%), Positives = 35/64 (54%)
Frame = +3
Query: 2 DQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDE 61
D EL ++D + +G I+ EL L+ LG S D +MI +D +GDG V+ +E
Sbjct: 306 DSKELRDACDLYDLDKNGLISANELHSVLKKLGEKCSLGDCRKMISNVDADGDGNVNFEE 485
Query: 62 FGEL 65
F ++
Sbjct: 486 FKKM 497
>TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regulated
calmodulin) (Calmodulin TACAM1-1) (Calmodulin TACAM1-2)
(Calmodulin TACAM1-3) (Calmodulin TACAM3-1) (Calmodulin
TACAM3-2) (Calmodulin TACAM3-3) (Calmodulin TACAM4-1)
(CaM protein), complete
Length = 615
Score = 119 bits (297), Expect = 3e-28
Identities = 62/139 (44%), Positives = 91/139 (64%)
Frame = +3
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 102 EFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 281
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++ +GFIS EL V+++LG K T E+ MI++
Sbjct: 282 LMARKMKDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 455
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG +N++EF ++M A
Sbjct: 456 DVDGDGQINYEEFVKVMMA 512
Score = 59.7 bits (143), Expect = 2e-10
Identities = 29/68 (42%), Positives = 46/68 (67%)
Frame = +3
Query: 74 DEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVN 133
D+ + KEAF++FD++GDG I+ +EL V+ SLG T + ++MI +VD DG+G ++
Sbjct: 90 DQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTID 263
Query: 134 FKEFQQMM 141
F EF +M
Sbjct: 264 FPEFLNLM 287
>TC19387 weakly similar to UP|Q9FYK2 (Q9FYK2) F21J9.28, partial (45%)
Length = 631
Score = 115 bits (288), Expect = 3e-27
Identities = 59/137 (43%), Positives = 93/137 (67%)
Frame = +1
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E+ IF+ FD+NGDG+I+ EL D + LG + E++ +M+ ++D NGDG++D+ EFGE
Sbjct: 46 EVREIFKKFDRNGDGKISCSELKDLMAALGSKTTAEEVRRMMAELDRNGDGHIDLKEFGE 225
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
++ ++++EAF ++D + +G IS +EL +V+ LG K +L DC+ MI V
Sbjct: 226 FHRG--GGGGNSKEIREAFEMYDLDKNGLISAKELHSVMKKLGEK--CSLGDCRKMIGNV 393
Query: 125 DVDGDGMVNFKEFQQMM 141
DVDGDG VNF+EF++MM
Sbjct: 394 DVDGDGNVNFEEFKKMM 444
Score = 56.2 bits (134), Expect = 2e-09
Identities = 31/84 (36%), Positives = 49/84 (57%)
Frame = +1
Query: 61 EFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
EFG + D E ++E F FD+NGDG IS EL ++++LG K T E+ + M
Sbjct: 1 EFGTSFFFFFSAMDNE--VREIFKKFDRNGDGKISCSELKDLMAALGSK--TTAEEVRRM 168
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAG 144
+ ++D +GDG ++ KEF + + G
Sbjct: 169 MAELDRNGDGHIDLKEFGEFHRGG 240
>TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(85%)
Length = 786
Score = 106 bits (265), Expect = 2e-24
Identities = 56/140 (40%), Positives = 89/140 (63%)
Frame = +3
Query: 2 DQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDE 61
D EL R+F FD NGDG+I+ EL + L++LG + E+L +++ +D + DG++++ E
Sbjct: 141 DMEELKRVFTRFDTNGDGKISVNELDNVLRSLGSGVPPEELKRVMVDLDGDHDGFINLSE 320
Query: 62 FGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMI 121
F ++ + E + +AF ++DQ+ +G IS EL VL+ LG+ + E+C MI
Sbjct: 321 FAAFCRSDTADGGASE-LHDAFELYDQDKNGLISAAELCKVLNRLGMNCSE--EECHKMI 491
Query: 122 KKVDVDGDGMVNFKEFQQMM 141
K VD DGDG VNF+EF++MM
Sbjct: 492 KSVDSDGDGNVNFEEFKKMM 551
Score = 52.4 bits (124), Expect = 3e-08
Identities = 29/64 (45%), Positives = 37/64 (57%)
Frame = +3
Query: 74 DEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVN 133
D+ E++K F FD NGDG IS EL VL SLG G E+ K ++ +D D DG +N
Sbjct: 138 DDMEELKRVFTRFDTNGDGKISVNELDNVLRSLG--SGVPPEELKRVMVDLDGDHDGFIN 311
Query: 134 FKEF 137
EF
Sbjct: 312 LSEF 323
>TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13E7_5
{Arabidopsis thaliana;}, partial (88%)
Length = 868
Score = 105 bits (261), Expect = 4e-24
Identities = 55/145 (37%), Positives = 89/145 (60%), Gaps = 3/145 (2%)
Frame = +1
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEF-G 63
EL IF+ FD+N DG +T+ ELS L++LG+ S E + I+K D N +G ++ EF
Sbjct: 214 ELREIFRSFDRNNDGSLTQLELSSLLRSLGLKPSAEQVEGFIQKADTNSNGLIEFSEFVA 393
Query: 64 ELYQTIMDEKD--EEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMI 121
+ ++ + EE +++ F VFD++G+GFI+ EL+ ++ LG H + E+ MI
Sbjct: 394 VVAPELLPARSPYTEEQLRQLFRVFDRDGNGFITAAELAHSMAKLG--HALSAEELTGMI 567
Query: 122 KKVDVDGDGMVNFKEFQQMMKAGAF 146
K+ D DGDGM++F+EF + + AF
Sbjct: 568 KEADADGDGMISFQEFAHAISSAAF 642
Score = 52.8 bits (125), Expect = 3e-08
Identities = 30/79 (37%), Positives = 49/79 (61%), Gaps = 7/79 (8%)
Frame = +1
Query: 66 YQTIMDEK-----DEEE--DMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCK 118
++ IM +K DEE+ +++E F FD+N DG ++ ELS++L SLGLK + E +
Sbjct: 157 HKEIMSKKQAVNLDEEQIAELREIFRSFDRNNDGSLTQLELSSLLRSLGLK--PSAEQVE 330
Query: 119 NMIKKVDVDGDGMVNFKEF 137
I+K D + +G++ F EF
Sbjct: 331 GFIQKADTNSNGLIEFSEF 387
>TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus annuus;} ,
partial (83%)
Length = 836
Score = 103 bits (256), Expect = 2e-23
Identities = 55/125 (44%), Positives = 82/125 (65%)
Frame = +2
Query: 19 GRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGELYQTIMDEKDEEED 78
G IT KEL +++LG +E +L MI ++D +G+G +D EF L M + D EE+
Sbjct: 164 GCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSEEE 343
Query: 79 MKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVNFKEFQ 138
+KEAF VFD++ +GFIS EL V+++LG K T E+ MI++ DVDGDG +N++EF
Sbjct: 344 LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIREADVDGDGQINYEEFV 517
Query: 139 QMMKA 143
++M A
Sbjct: 518 KVMMA 532
Score = 51.2 bits (121), Expect = 8e-08
Identities = 28/88 (31%), Positives = 50/88 (56%), Gaps = 9/88 (10%)
Frame = +2
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
EL F++FDK+ +G I+ EL + NLG +++E++ +MI + DV+GDG ++ +EF +
Sbjct: 341 ELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEFVK 520
Query: 65 L---------YQTIMDEKDEEEDMKEAF 83
+ Y I+ E+ D + F
Sbjct: 521 VMMAK*GPFNYNYILKTYKEKRDRADKF 604
>TC8061 UP|AAA34013 (AAA34013) Calmodulin, complete
Length = 568
Score = 51.6 bits (122), Expect(3) = 1e-19
Identities = 26/59 (44%), Positives = 41/59 (69%)
Frame = +2
Query: 85 VFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVNFKEFQQMMKA 143
+ D++ +GFIS EL V+++LG K T E+ MI++ DVDGDG +N++EF ++M A
Sbjct: 320 LLDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIREADVDGDGQINYEEFVKVMMA 490
Score = 43.9 bits (102), Expect(3) = 1e-19
Identities = 20/49 (40%), Positives = 29/49 (58%)
Frame = +1
Query: 45 MIEKIDVNGDGYVDIDEFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGF 93
MI ++D +G+G +D EF L M + D EE++KEAF V Q + F
Sbjct: 199 MINEVDADGNGTIDFPEFLNLMARKMKDTDSEEELKEAFRVVGQGSERF 345
Score = 43.5 bits (101), Expect = 2e-05
Identities = 19/55 (34%), Positives = 37/55 (66%)
Frame = +2
Query: 8 RIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEF 62
++ + DK+ +G I+ EL + NLG +++E++ +MI + DV+GDG ++ +EF
Sbjct: 308 KLSALLDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEF 472
Score = 38.1 bits (87), Expect = 7e-04
Identities = 16/34 (47%), Positives = 26/34 (76%)
Frame = +3
Query: 74 DEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLG 107
++ + KEAF++FD++GDG I+ +EL V+ SLG
Sbjct: 66 EQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG 167
Score = 35.0 bits (79), Expect(3) = 1e-19
Identities = 17/38 (44%), Positives = 24/38 (62%)
Frame = +3
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDL 42
E F +FDK+GDG IT KEL +++LG +E +L
Sbjct: 78 EFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAEL 191
>TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodulin-like
protein {Arabidopsis thaliana;} , partial (72%)
Length = 1174
Score = 87.4 bits (215), Expect = 1e-18
Identities = 47/134 (35%), Positives = 85/134 (63%), Gaps = 1/134 (0%)
Frame = +3
Query: 10 FQMFDKNGDGRITKKELSDSLQNLG-ICISEEDLVQMIEKIDVNGDGYVDIDEFGELYQT 68
F++ D++ DG +++ +L L LG + +S+E++ M+ ++D +G G + ++ L +
Sbjct: 306 FRLIDRDNDGVVSRADLVAVLTRLGALQLSDEEVEVMLREVDGDGRGSISVEA---LMER 476
Query: 69 IMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDG 128
+ D E +++EAF VFD + DG IS EEL V +++G TL++C+ MI+ VD +G
Sbjct: 477 VGPGSDPETELREAFEVFDTDRDGRISAEELLRVFTAIG-DERCTLDECRRMIEGVDRNG 653
Query: 129 DGMVNFKEFQQMMK 142
DG V F++F +MM+
Sbjct: 654 DGFVCFEDFSRMME 695
Score = 42.0 bits (97), Expect = 5e-05
Identities = 24/65 (36%), Positives = 38/65 (57%), Gaps = 2/65 (3%)
Frame = +3
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLG--ICISEEDLVQMIEKIDVNGDGYVDIDEF 62
EL F++FD + DGRI+ +EL +G C +E +MIE +D NGDG+V ++F
Sbjct: 504 ELREAFEVFDTDRDGRISAEELLRVFTAIGDERCTLDE-CRRMIEGVDRNGDGFVCFEDF 680
Query: 63 GELYQ 67
+ +
Sbjct: 681 SRMME 695
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 82.8 bits (203), Expect = 2e-17
Identities = 46/138 (33%), Positives = 84/138 (60%), Gaps = 2/138 (1%)
Frame = +1
Query: 9 IFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGELYQT 68
+F + D + DGR++ +EL L+ +G ++++++ ++E DV+G+G +D EF + T
Sbjct: 28 MFTLMDTDKDGRVSYEELKAGLRKVGSQLADQEIKMLMEVADVDGNGVLDYGEF--VAVT 201
Query: 69 I-MDEKDEEEDMKEAFNVFDQNGDGFI-SGEELSAVLSSLGLKHGKTLEDCKNMIKKVDV 126
I + + + +E +AF FD++G G+I SGE A+ G+ L D ++++VD
Sbjct: 202 IHLQKMENDEHFHKAFKFFDKDGSGYIESGELQEALADESGVTDADVLND---IMREVDT 372
Query: 127 DGDGMVNFKEFQQMMKAG 144
D DG ++++EF MMKAG
Sbjct: 373 DKDGRISYEEFVAMMKAG 426
>CB828304
Length = 489
Score = 67.8 bits (164), Expect = 8e-13
Identities = 41/128 (32%), Positives = 70/128 (54%), Gaps = 7/128 (5%)
Frame = +1
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
+L IF FD + DG +T EL+ L++LG+ S + L ++ +D NG+G V EF E
Sbjct: 118 QLREIFARFDMDSDGSLTMLELAALLRSLGLKPSGDQLHDLLSNMDSNGNGSV---EFDE 288
Query: 65 LYQTIMDEKDE-------EEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDC 117
L +TI+ + +E + + F FD++ +GFIS EL+ ++ +G T ++
Sbjct: 289 LVRTILPDLKNNAEVLLNQEQLLDVFKCFDRDSNGFISAAELAGAMAKMG--QPLTYKEL 462
Query: 118 KNMIKKVD 125
MI++ D
Sbjct: 463 TEMIREAD 486
>TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 817
Score = 62.0 bits (149), Expect = 4e-11
Identities = 39/134 (29%), Positives = 70/134 (52%)
Frame = +3
Query: 10 FQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGELYQTI 69
F +FD +GDG+I EL +++LG ++ L ++ + ++ D F +L
Sbjct: 153 FTLFDTDGDGKIAPSELGILMRSLGGNPTQAQLKSIVAEENLTSP--FDFPRFLDLMAKH 326
Query: 70 MDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGD 129
M + + +++AF V D++ GF+S EL +L+S+G K D I++VDV D
Sbjct: 327 MKPEPFDRQLRDAFKVIDKDSTGFVSVTELRHILTSIGEKLEPAEFD--EWIREVDVGSD 500
Query: 130 GMVNFKEFQQMMKA 143
G + +++F M A
Sbjct: 501 GKIRYEDFIARMVA 542
Score = 38.1 bits (87), Expect = 7e-04
Identities = 20/55 (36%), Positives = 32/55 (57%), Gaps = 3/55 (5%)
Frame = +3
Query: 56 YVDIDEFGELYQTIMDEK---DEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLG 107
+V + + + +T M + D+ MKEAF +FD +GDG I+ EL ++ SLG
Sbjct: 63 FVALSQHKRIIETKMGKDLSDDQVSSMKEAFTLFDTDGDGKIAPSELGILMRSLG 227
>CB828334
Length = 541
Score = 61.6 bits (148), Expect = 6e-11
Identities = 40/152 (26%), Positives = 69/152 (45%), Gaps = 18/152 (11%)
Frame = +2
Query: 4 GELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFG 63
G+ IF+ FD++ +G I ++EL L I +EE++ + E D+N D ++ EF
Sbjct: 47 GKCKAIFKQFDEDSNGAIDQEELKKCFSKLEISFTEEEVNDLFEACDINEDMGMEFSEFI 226
Query: 64 ELYQTIMDEKDEE------------------EDMKEAFNVFDQNGDGFISGEELSAVLSS 105
L + KD+ E + + F D+N DG++S E+ ++
Sbjct: 227 VLLCLVHLLKDDRAAVQAKSRIGMPDLETTFETLVDTFVFLDKNKDGYVSKSEMVQAINE 406
Query: 106 LGLKHGKTLEDCKNMIKKVDVDGDGMVNFKEF 137
+ +++D D +GMVNFKEF
Sbjct: 407 TVSGERSSGRIAMKRFEEMDWDKNGMVNFKEF 502
>AU089222
Length = 345
Score = 59.3 bits (142), Expect = 3e-10
Identities = 25/61 (40%), Positives = 45/61 (72%)
Frame = +3
Query: 2 DQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDE 61
D + RIF+ FD NGDG+I+ EL ++L+ LG ++ E++ +M+++ID++GDG++ +E
Sbjct: 66 DVADRERIFKRFDANGDGQISSSELGEALKTLG-SVTAEEVQRMMDEIDIDGDGFISYEE 242
Query: 62 F 62
F
Sbjct: 243 F 245
Score = 57.4 bits (137), Expect = 1e-09
Identities = 27/73 (36%), Positives = 42/73 (56%)
Frame = +3
Query: 71 DEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDG 130
D+ + D + F FD NGDG IS EL L +LG T E+ + M+ ++D+DGDG
Sbjct: 54 DDPQDVADRERIFKRFDANGDGQISSSELGEALKTLG---SVTAEEVQRMMDEIDIDGDG 224
Query: 131 MVNFKEFQQMMKA 143
++++EF +A
Sbjct: 225 FISYEEFTSFARA 263
>CB828195
Length = 551
Score = 58.5 bits (140), Expect = 5e-10
Identities = 29/62 (46%), Positives = 39/62 (62%)
Frame = +3
Query: 64 ELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKK 123
EL + D++ E++K+AF+VFD+N DGFI EL VL LGLK +E CK MI+
Sbjct: 363 ELSELFEDQEPSLEEVKQAFDVFDENRDGFIDARELHRVLCVLGLKEEAGIEKCKIMIRN 542
Query: 124 VD 125
D
Sbjct: 543 FD 548
>TC18670 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(41%)
Length = 329
Score = 54.3 bits (129), Expect = 9e-09
Identities = 24/58 (41%), Positives = 37/58 (63%)
Frame = +3
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEF 62
EL R+F FD N DG+I+ EL L+ LG + E+L Q++E +D + DG++ + EF
Sbjct: 114 ELERVFNRFDANSDGKISVDELDSVLRTLGSGVPPEELQQVMEDLDTDRDGFISLPEF 287
>TC13869 weakly similar to UP|Q9FR00 (Q9FR00) Avr9/Cf-9 rapidly elicited
protein 31, partial (45%)
Length = 547
Score = 54.3 bits (129), Expect = 9e-09
Identities = 33/86 (38%), Positives = 48/86 (55%), Gaps = 8/86 (9%)
Frame = +3
Query: 6 LARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGEL 65
L RIF MFDKNGD IT +E+S +L LG+ ++ MI G+ + D+F L
Sbjct: 288 LRRIFDMFDKNGDCMITVEEISQALNLLGLEAEVAEIDSMIRSYIRPGNEGLTYDDFMAL 467
Query: 66 YQTIMD--------EKDEEEDMKEAF 83
+++I D EK +E D++EAF
Sbjct: 468 HESIGDTFFGFVEEEKGDESDLREAF 545
Score = 34.3 bits (77), Expect = 0.010
Identities = 18/62 (29%), Positives = 36/62 (58%)
Frame = +3
Query: 79 MKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVNFKEFQ 138
++ F++FD+NGD I+ EE+S L+ LGL+ + + +MI+ G+ + + +F
Sbjct: 288 LRRIFDMFDKNGDCMITVEEISQALNLLGLE--AEVAEIDSMIRSYIRPGNEGLTYDDFM 461
Query: 139 QM 140
+
Sbjct: 462 AL 467
>TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partial (19%)
Length = 657
Score = 53.1 bits (126), Expect = 2e-08
Identities = 32/96 (33%), Positives = 53/96 (54%), Gaps = 5/96 (5%)
Frame = +2
Query: 54 DGYVDIDEFGELYQTIMDEKDEEED-MKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGK 112
DG +D EF + T+ K E +D + AF FD++ GFI+ +EL + G+
Sbjct: 14 DGTIDYIEF--VTATMHRHKLERDDHLN*AFPYFDKDSSGFITRDELEIAMKEYGMGDDA 187
Query: 113 TLE----DCKNMIKKVDVDGDGMVNFKEFQQMMKAG 144
T++ + +I +VD D DG +N++EF MM++G
Sbjct: 188 TIKEIISEVDTIISEVDTDHDGRINYEEFCAMMRSG 295
>CB828250
Length = 485
Score = 49.3 bits (116), Expect = 3e-07
Identities = 33/118 (27%), Positives = 62/118 (51%), Gaps = 5/118 (4%)
Frame = +1
Query: 10 FQMFDKNGDGRITKKELSDSLQNL--GICISEEDLVQMIEKIDVNGDGYVDIDEFGELYQ 67
F + D + DG+I++ +L + G+ ++ + M+ D + DG+V+ +EF +
Sbjct: 130 FDVLDADCDGKISRDDLRAFYAGIYGGVASGDDVIGAMMTVADTDKDGFVEYEEF----E 297
Query: 68 TIMDEKDEE---EDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIK 122
++ E ++ M+E F V D++GDG +S +L + ++S G T ED MIK
Sbjct: 298 RVVTENNKPLGCGAMEEVFKVMDKDGDGKLSHHDLKSYMASAGF--SATDEDINAMIK 465
Score = 35.8 bits (81), Expect = 0.003
Identities = 13/44 (29%), Positives = 28/44 (63%)
Frame = +1
Query: 4 GELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIE 47
G + +F++ DK+GDG+++ +L + + G ++ED+ MI+
Sbjct: 334 GAMEEVFKVMDKDGDGKLSHHDLKSYMASAGFSATDEDINAMIK 465
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.137 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,687,625
Number of Sequences: 28460
Number of extensions: 16388
Number of successful extensions: 167
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 132
length of query: 152
length of database: 4,897,600
effective HSP length: 82
effective length of query: 70
effective length of database: 2,563,880
effective search space: 179471600
effective search space used: 179471600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 51 (24.3 bits)
Medicago: description of AC122169.9


