

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.2 + phase: 0
(395 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
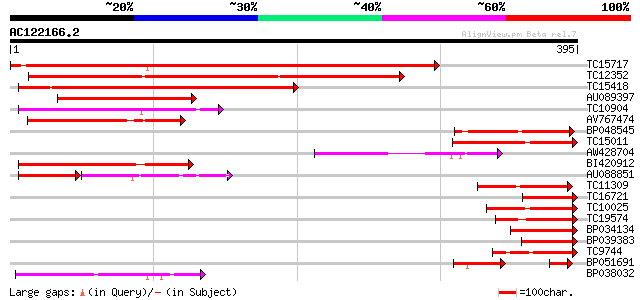
Score E
Sequences producing significant alignments: (bits) Value
TC15717 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 282 6e-77
TC12352 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 232 9e-62
TC15418 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 224 1e-59
AU089397 114 3e-26
TC10904 weakly similar to UP|Q62258 (Q62258) Spi2 proteinase inh... 107 3e-24
AV767474 100 4e-22
BP048545 99 2e-21
TC15011 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragm... 97 4e-21
AW428704 94 4e-20
BI420912 91 4e-19
AU088851 52 7e-15
TC11309 similar to UP|Q9FUV8 (Q9FUV8) Phloem serpin-1, partial (... 73 7e-14
TC16721 weakly similar to UP|P93692 (P93692) Serpin, partial (9%) 71 3e-13
TC10025 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 66 1e-11
TC19574 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 65 3e-11
BP034134 64 3e-11
BP039383 63 1e-10
TC9744 similar to UP|Q40066 (Q40066) Protein zx, partial (7%) 61 4e-10
BP051691 47 2e-08
BP038032 55 3e-08
>TC15717 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (26%)
Length = 925
Score = 282 bits (722), Expect = 6e-77
Identities = 152/303 (50%), Positives = 206/303 (67%), Gaps = 4/303 (1%)
Frame = +1
Query: 1 MQNLIGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQL 60
+Q I KS +++++ L S + EN+VFSPLSL+ L+++A G+ G T +L
Sbjct: 25 LQKSISKS---QEDVALSFANRLFSTEAYHNENIVFSPLSLHVALAIMAAGAHGSTLDEL 195
Query: 61 LSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPC---LSYVNGVWVEQTIPLQPSFKQLM 117
LSFL+ +S L ++ SQ+VS+V SD A P LS+ NG+WV++++ L SFKQL+
Sbjct: 196 LSFLRFDSVDHLNTIFSQVVSAVFSDNDDAAPPPTHRLSFANGMWVDKSLSLTHSFKQLV 375
Query: 118 NTDFKAAFAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFK 177
T +KA +VDF NKA++V +EVN W EK T GLIK LL PG+V+ TRLIFANAL+FK
Sbjct: 376 ATHYKATLDSVDFWNKADQVCDEVNLWVEKGTNGLIKELLSPGAVDKTTRLIFANALHFK 555
Query: 178 GVWKQQFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDE-RA 236
G W+ +F + Y F LL+G SV VP MT+ +Q I DGFK+LGLPYKQG DE R
Sbjct: 556 GEWEHKFLARYSYSYRFHLLDGTSVVVPLMTNDEEQLIRVFDGFKILGLPYKQGTDEKRL 735
Query: 237 FSIYFFLPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLK 296
FS+Y LP KDGLS+LI K+AS+ FLE LP++KV++ F IPRF+ISF EAS++LK
Sbjct: 736 FSMYILLPHAKDGLSDLIRKMASEPGFLEGKLPQQKVKLNFFLIPRFDISFAFEASDVLK 915
Query: 297 KLG 299
+ G
Sbjct: 916 EFG 924
>TC12352 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (25%)
Length = 799
Score = 232 bits (591), Expect = 9e-62
Identities = 126/264 (47%), Positives = 181/264 (67%), Gaps = 2/264 (0%)
Frame = +1
Query: 14 NISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQSESTGDLK 73
++ ++ T+HL S + +E+N+++SPLSL LS+IA GSEG T +LLSFL+ +S +L
Sbjct: 16 DVPLSFTQHLFSKEDYQEKNLIYSPLSLYAALSVIAAGSEGRTFDELLSFLRFDSIDNLN 195
Query: 74 SLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFAAVDFVNK 133
+ SQ +S V D A L + NG++++ T+ L F++L++T + A ++DF +
Sbjct: 196 TFFSQAISPVFFDNDAASP--LQHYNGIFIDTTVSLSYPFRRLLSTHYNANLTSLDFNLR 369
Query: 134 ANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDTTKTKDYD 193
+V E+N E++T G I LLPPG+V +LTRLIFANAL F+G+WK +FD T
Sbjct: 370 GGKVLHEMNSLIEEDTNGHITQLLPPGTVTNLTRLIFANALCFQGMWKHKFDGL-TYVSP 546
Query: 194 FDLLNGKSVKVPFMTS-KNDQFISSLDGFKVLGLPYKQGKD-ERAFSIYFFLPDKKDGLS 251
F+LLNG SVKVPFMT+ KN Q++ + DGFK+L LPYKQG+D +R FS+ FLPD +DGLS
Sbjct: 547 FNLLNGTSVKVPFMTTCKNTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLS 726
Query: 252 NLIDKVASDSEFLERNLPRRKVEV 275
LI K++S+ FL+ LPRRKV V
Sbjct: 727 ALIQKLSSEPCFLKGKLPRRKVRV 798
>TC15418 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (44%)
Length = 602
Score = 224 bits (572), Expect = 1e-59
Identities = 116/195 (59%), Positives = 143/195 (72%)
Frame = +1
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQS 66
+S T++S+ I K L S K++N VFSPLSL+ VLS++A GSEGPT QLL+FL+S
Sbjct: 19 ESIETQTDVSLTIAKLLFSKHS-KDKNAVFSPLSLHVVLSILAAGSEGPTLHQLLTFLRS 195
Query: 67 ESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFA 126
ST L S SQLVS VLSD + GGP L + +GVWVEQ++ L PSFKQL+ D+ AA A
Sbjct: 196 NSTDHLNSFASQLVSVVLSDASSVGGPRLCFADGVWVEQSLTLNPSFKQLVTADYHAALA 375
Query: 127 AVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDT 186
+VDF KA EV EVN WAEKET GL+K+LLP GSV++ TRLIFANALYFKG W ++FD
Sbjct: 376 SVDFQTKAVEVANEVNAWAEKETNGLVKDLLPAGSVDASTRLIFANALYFKGAWTEKFDA 555
Query: 187 TKTKDYDFDLLNGKS 201
+ TKD DF LL+G S
Sbjct: 556 SMTKDCDFHLLDGTS 600
>AU089397
Length = 306
Score = 114 bits (285), Expect = 3e-26
Identities = 57/97 (58%), Positives = 71/97 (72%)
Frame = +3
Query: 34 VVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGP 93
+VFSPLSL VLS+IA GS+G T +LLSFL S ST L S SQL+S+VLSD APAGGP
Sbjct: 9 IVFSPLSLQVVLSIIAAGSDGSTLDELLSFLGSNSTDHLNSFASQLISTVLSDAAPAGGP 188
Query: 94 CLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFAAVDF 130
L + VWVE+++P+ SF Q+MNTD+ A +VDF
Sbjct: 189 RLCFAXXVWVEKSLPVNHSFXQVMNTDYXATLTSVDF 299
>TC10904 weakly similar to UP|Q62258 (Q62258) Spi2 proteinase inhibitor,
partial (8%)
Length = 576
Score = 107 bits (267), Expect = 3e-24
Identities = 60/148 (40%), Positives = 87/148 (58%), Gaps = 5/148 (3%)
Frame = -1
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQS 66
KS + ++++ +TKHL S + +N+VFSP SL LS++A GS+ T +LLSFL+
Sbjct: 441 KSKSKSMDVALGLTKHLFSKADYQGKNIVFSPFSLQAALSVMAAGSKDRTLDELLSFLRF 262
Query: 67 ESTGDLKSLCSQLVSSVLSDGAPA-----GGPCLSYVNGVWVEQTIPLQPSFKQLMNTDF 121
+S DL + SQ++ VL A A G L + NG+W + ++ L FKQL+ T +
Sbjct: 261 DSIDDLTTFFSQVIFPVLISDAAADADTDGSHHLCFANGIWADDSLSLSHRFKQLVATHY 82
Query: 122 KAAFAAVDFVNKANEVGEEVNFWAEKET 149
KA A+DF + EV EVN W EKET
Sbjct: 81 KATLTALDF--QTTEVHREVNSWIEKET 4
>AV767474
Length = 380
Score = 100 bits (249), Expect = 4e-22
Identities = 51/110 (46%), Positives = 80/110 (72%)
Frame = +3
Query: 13 TNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQSESTGDL 72
T++++ ITKHL S Q+ +++N++FSPLSL+ VLS++A GS G T +LLSFL+ +S L
Sbjct: 63 TDVALTITKHLFSKQEYQDKNLMFSPLSLHAVLSLLAAGSAGSTLDELLSFLRFDSIDHL 242
Query: 73 KSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFK 122
+ SQL+S+ AP+ P L++VNG+WV++++ L SFKQL+ T +K
Sbjct: 243 NTFFSQLISA----AAPSSHP-LTFVNGMWVDKSVSLSNSFKQLLATHYK 377
>BP048545
Length = 452
Score = 98.6 bits (244), Expect = 2e-21
Identities = 52/83 (62%), Positives = 61/83 (72%)
Frame = -2
Query: 311 KMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHP 370
KMVDS +LYVS IF KSFIEVNEEGT+AAA + + RS P P +DFVADHP
Sbjct: 451 KMVDS----DLYVSNIFHKSFIEVNEEGTRAAAASAGIVLKRSL--SPRPTRVDFVADHP 290
Query: 371 FLFLIREEFSGTILFVGKVVNPL 393
F F+IREE +GTILF G+V+NPL
Sbjct: 289 FFFVIREEMTGTILFTGQVLNPL 221
>TC15011 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (20%)
Length = 623
Score = 97.4 bits (241), Expect = 4e-21
Identities = 54/88 (61%), Positives = 64/88 (72%), Gaps = 1/88 (1%)
Frame = +2
Query: 309 LTKMVDSP-ISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVA 367
LTKMVDS + Q LYVS IF KSFIEVNEEGT+AAA + I RS +DFVA
Sbjct: 17 LTKMVDSSTVGQGLYVSNIFHKSFIEVNEEGTEAAAASAVTIKLRSLQVLTR---VDFVA 187
Query: 368 DHPFLFLIREEFSGTILFVGKVVNPLDG 395
DHPFLF+IRE+ +GT+LF G+V+NPL G
Sbjct: 188 DHPFLFVIREDLTGTVLFTGQVLNPLAG 271
>AW428704
Length = 369
Score = 94.0 bits (232), Expect = 4e-20
Identities = 63/136 (46%), Positives = 79/136 (57%), Gaps = 5/136 (3%)
Frame = +1
Query: 213 QFISSLDGFKVLGLPYKQGKDE-RAFSIYFFLPDKKDGLSNLIDKVASDSEFLERNLPRR 271
+ I DGFK+LGLPYKQG DE R FS+Y LP KDGLS+LI K+AS+ FL
Sbjct: 13 RLIRVFDGFKILGLPYKQGTDEKRLFSMYILLPHAKDGLSDLIRKMASEPGFL------- 171
Query: 272 KVEVGKFRIPRFNISFEIEASELLKKLGLALPFTL--GGLTKM--VDSPISQELYVSGIF 327
EAS++LK+ G+ PF+ TKM V+SP+ L V IF
Sbjct: 172 ------------------EASDVLKEFGVVSPFSQRDADFTKMVKVNSPL-DALSVESIF 294
Query: 328 QKSFIEVNEEGTKAAA 343
QK FI+VNE+GT+AAA
Sbjct: 295 QKVFIKVNEQGTEAAA 342
>BI420912
Length = 508
Score = 90.5 bits (223), Expect = 4e-19
Identities = 52/123 (42%), Positives = 82/123 (66%), Gaps = 1/123 (0%)
Frame = +2
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQS 66
KS +N + ++ TKH+LS ++ +E+N+VFSPLSL LS++A G++G T +LLSFL+
Sbjct: 158 KSKSNSIDAVLSFTKHVLSKEEYQEKNLVFSPLSLYAALSVMAAGADGLTLDELLSFLRF 337
Query: 67 ESTGDLKSLCSQLVSSVL-SDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAF 125
+S L + SQ +S VL SD LS+ NG+W +Q++ + SFKQL++T +KA
Sbjct: 338 DSVDHLTTFFSQDLSPVLFSDDH------LSFANGMWGDQSLSIFHSFKQLVSTHYKATL 499
Query: 126 AAV 128
A++
Sbjct: 500 ASL 508
>AU088851
Length = 663
Score = 52.4 bits (124), Expect(2) = 7e-15
Identities = 43/108 (39%), Positives = 59/108 (53%), Gaps = 3/108 (2%)
Frame = +3
Query: 51 GSEGPTQKQLLSFLQSESTGDLKSLCSQLVSSVL--SDGAPAGGPCLSYVNGVWVEQTIP 108
GSEG T +LLSFL+ +ST L S SQLVS VL +D AP P LS+ + +W ++T+
Sbjct: 267 GSEGSTLDELLSFLRFDSTDRLNSFTSQLVSDVLYVNDAAPP-SPRLSFASRMWADKTLF 443
Query: 109 LQ-PSFKQLMNTDFKAAFAAVDFVNKANEVGEEVNFWAEKETKGLIKN 155
L PS+ + K A++DF K + V W K+ IKN
Sbjct: 444 LSLPSYNLWLL--IKELLASLDF-XKQGKKXCRV*IWVGKKXMS-IKN 575
Score = 44.3 bits (103), Expect(2) = 7e-15
Identities = 19/43 (44%), Positives = 34/43 (78%)
Frame = +1
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIA 49
+S ++ +++++ +TKHL S + + EN+VFSPLSL+ VLS++A
Sbjct: 133 ESTSHQSDVALTLTKHLFSKEVYQHENLVFSPLSLHVVLSIMA 261
>TC11309 similar to UP|Q9FUV8 (Q9FUV8) Phloem serpin-1, partial (10%)
Length = 472
Score = 73.2 bits (178), Expect = 7e-14
Identities = 36/66 (54%), Positives = 49/66 (73%)
Frame = +3
Query: 327 FQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFV 386
F + ++++EEGT AAA T ++SRS P IDFVADHPF FLIRE+F+GTILF+
Sbjct: 90 FTRLSLKLDEEGTTAAAATAMLLASRS--GPSVRAGIDFVADHPFFFLIREDFTGTILFI 263
Query: 387 GKVVNP 392
G+V++P
Sbjct: 264 GQVLHP 281
>TC16721 weakly similar to UP|P93692 (P93692) Serpin, partial (9%)
Length = 553
Score = 71.2 bits (173), Expect = 3e-13
Identities = 30/38 (78%), Positives = 36/38 (93%)
Frame = +2
Query: 358 PPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNPLDG 395
PPPP ++F+ADHPFLFLIRE+FSGTILFVG+V+NPL G
Sbjct: 20 PPPPGLEFIADHPFLFLIREDFSGTILFVGQVLNPLGG 133
>TC10025 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (15%)
Length = 672
Score = 65.9 bits (159), Expect = 1e-11
Identities = 36/63 (57%), Positives = 43/63 (68%)
Frame = +1
Query: 333 EVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
EVNEE T A A T + + P IDFVADHPFL L+RE+ +GTILFVG+V+NP
Sbjct: 1 EVNEEETVAVAATAARMVLGGAGLCPH---IDFVADHPFLLLVREDITGTILFVGQVLNP 171
Query: 393 LDG 395
LDG
Sbjct: 172 LDG 180
>TC19574 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (8%)
Length = 414
Score = 64.7 bits (156), Expect = 3e-11
Identities = 33/57 (57%), Positives = 40/57 (69%)
Frame = -3
Query: 339 TKAAAVTVSFISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNPLDG 395
TKA A S + + R P IDFVA+HPFLFLIREE +GT LF+G+V+NPLDG
Sbjct: 409 TKATAAAGSLLGTYFR-----PAGIDFVANHPFLFLIREELTGTFLFIGQVLNPLDG 254
>BP034134
Length = 556
Score = 64.3 bits (155), Expect = 3e-11
Identities = 29/46 (63%), Positives = 35/46 (76%)
Frame = -1
Query: 350 SSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNPLDG 395
+ R Y P IDF ADHPFLFLIRE+ +GTILF+G+V+NPLDG
Sbjct: 520 NERESYVSDRPTGIDFEADHPFLFLIREDLTGTILFIGQVLNPLDG 383
>BP039383
Length = 340
Score = 62.8 bits (151), Expect = 1e-10
Identities = 28/39 (71%), Positives = 32/39 (81%)
Frame = -1
Query: 357 PPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNPLDG 395
P P +DFVADHPFLF+IREE +GTILF G+VVNPL G
Sbjct: 322 PSPKTRVDFVADHPFLFVIREEMTGTILFTGQVVNPLAG 206
>TC9744 similar to UP|Q40066 (Q40066) Protein zx, partial (7%)
Length = 527
Score = 60.8 bits (146), Expect = 4e-10
Identities = 34/59 (57%), Positives = 43/59 (72%)
Frame = +1
Query: 337 EGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNPLDG 395
+GT+A + TV +S R R FVADHPFLFLIRE+F+GTILFVG+V+NPL+G
Sbjct: 1 KGTEATSATV--VSGRKRLGGFLGTY--FVADHPFLFLIREDFTGTILFVGQVLNPLEG 165
>BP051691
Length = 428
Score = 47.0 bits (110), Expect(2) = 2e-08
Identities = 24/40 (60%), Positives = 29/40 (72%), Gaps = 4/40 (10%)
Frame = -2
Query: 310 TKMVDSPI----SQELYVSGIFQKSFIEVNEEGTKAAAVT 345
T MVDSP S ELY+ +F K+FIEVNEEGT+AAA +
Sbjct: 406 TNMVDSPSDEFPSDELYIDNMFHKAFIEVNEEGTEAAAAS 287
Score = 27.7 bits (60), Expect(2) = 2e-08
Identities = 10/16 (62%), Positives = 15/16 (93%)
Frame = -1
Query: 377 EEFSGTILFVGKVVNP 392
E+ SGTILF+G+V++P
Sbjct: 275 EDLSGTILFIGRVLHP 228
>BP038032
Length = 437
Score = 54.7 bits (130), Expect = 3e-08
Identities = 40/141 (28%), Positives = 72/141 (50%), Gaps = 9/141 (6%)
Frame = +1
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFL 64
+ KS T++++++TKHL S ++ +++N++FSPLSL+ LS++A GS G T + +FL
Sbjct: 22 VEKSMRCQTDVALSVTKHLFSKEEYQQKNLIFSPLSLHVALSVMAGGSAGGTLSR--TFL 195
Query: 65 QSESTGDLKSLCSQLVSSVLSDGAPAGGPC------LSYVNGVWVE---QTIPLQPSFKQ 115
G + L + +L+ P L NG+WV+ P+ P+
Sbjct: 196 SFFGIGSHWKNFNNLSNLILNFHRGPWTPVVISPLRLPLANGMWVD*ITFHFPILPN--N 369
Query: 116 LMNTDFKAAFAAVDFVNKANE 136
+KA A++DF ++
Sbjct: 370 W*PLHYKATLASLDFQKNGDQ 432
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,144,224
Number of Sequences: 28460
Number of extensions: 86583
Number of successful extensions: 1954
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 1276
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1771
length of query: 395
length of database: 4,897,600
effective HSP length: 92
effective length of query: 303
effective length of database: 2,279,280
effective search space: 690621840
effective search space used: 690621840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC122166.2


