

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122165.6 + phase: 0
(424 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
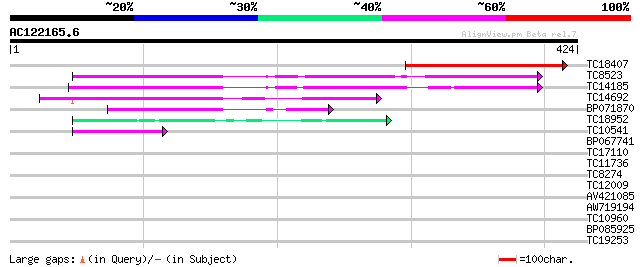
Score E
Sequences producing significant alignments: (bits) Value
TC18407 similar to UP|Q9LG24 (Q9LG24) F14J16.13, partial (28%) 236 7e-63
TC8523 homologue to UP|Q8W1A0 (Q8W1A0) Cysteine synthase, complete 145 1e-35
TC14185 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthase... 143 6e-35
TC14692 similar to UP|CYSK_CITLA (Q43317) Cysteine synthase (Be... 130 3e-31
BP071870 67 4e-12
TC18952 similar to GB|AAD54324.1|5853119|AF177212 threonine dehy... 46 1e-05
TC10541 similar to UP|CYSL_CAPAN (P31300) Cysteine synthase, chl... 45 2e-05
BP067741 35 0.023
TC17110 similar to UP|Q9FS26 (Q9FS26) Plastidic cysteine synthas... 35 0.031
TC11736 31 0.44
TC8274 similar to GB|AAA36350.1|189049|HUMNADH NADH dehydrogenas... 28 2.2
TC12009 similar to PIR|T52589|T52589 ribose-phosphate diphosphok... 28 2.9
AV421085 28 3.7
AW719194 27 6.4
TC10960 similar to PIR|B96700|B96700 protein F12A21.17 [imported... 27 6.4
BP085925 27 8.3
TC19253 27 8.3
>TC18407 similar to UP|Q9LG24 (Q9LG24) F14J16.13, partial (28%)
Length = 380
Score = 236 bits (601), Expect = 7e-63
Identities = 115/121 (95%), Positives = 120/121 (99%)
Frame = +3
Query: 297 GVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAVEMARFLVKN 356
GVMYTKEEAEGRRLKNPFDTITEGIGINRVT+NFA AKLDGAFRGTDMEAVEMARFL+KN
Sbjct: 6 GVMYTKEEAEGRRLKNPFDTITEGIGINRVTRNFAMAKLDGAFRGTDMEAVEMARFLLKN 185
Query: 357 DGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEYLSQLGLTPKAT 416
DGLFLGSSSAMNCVGAVR+AQ++GPGHTIVTILCDSGMRHLSKFYNAEYLSQLGLTPKAT
Sbjct: 186 DGLFLGSSSAMNCVGAVRVAQTLGPGHTIVTILCDSGMRHLSKFYNAEYLSQLGLTPKAT 365
Query: 417 G 417
G
Sbjct: 366 G 368
>TC8523 homologue to UP|Q8W1A0 (Q8W1A0) Cysteine synthase, complete
Length = 1361
Score = 145 bits (366), Expect = 1e-35
Identities = 107/353 (30%), Positives = 157/353 (44%), Gaps = 2/353 (0%)
Frame = +3
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG-G 106
+ IG TPL+ +N L+D + K E + P SVKDR+ +I +A E G + G
Sbjct: 111 VTQLIGKTPLVYLNKLADGCVARVAAKLELMEPCSSVKDRIGYSMIADAEEKGLITPGKS 290
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
++ E ++G+T I +A +A A G K + +P ++E+ I+ A GA + P
Sbjct: 291 VLIEPTSGNTGIGLAFMAAAKGYKLIITMPASMSLERRTILLAFGAELVLTDPA------ 452
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
KG + G + PN + QF
Sbjct: 453 -------------------------KG-----MKGAVQKAEEIVAKTPNS----YMLQQF 530
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPP 286
EN AN + HYE TGPEIW+ T G +DA V+ GTGGT+ G K+L+E+N NIK ++P
Sbjct: 531 ENAANPKVHYETTGPEIWKGTAGKVDALVSGVGTGGTITGAGKYLKEQNSNIKMIAVEPV 710
Query: 287 GSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEA 346
S V+ G P +GIG V LD + + EA
Sbjct: 711 ES----PVLSG-------------GKPGPHKIQGIGAGFVPGVLDVNLLDEVIQISSDEA 839
Query: 347 VEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
+E A+ L +GLF+G SS A++IA + G IV + G R+LS
Sbjct: 840 IETAKLLALKEGLFVGISSGAAAAAAIKIAKRPENAGKLIVVVFPSFGERYLS 998
>TC14185 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthase, partial
(96%)
Length = 1506
Score = 143 bits (360), Expect = 6e-35
Identities = 108/356 (30%), Positives = 159/356 (44%), Gaps = 2/356 (0%)
Frame = +1
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
K + + IG TPL+ +N +++ G I K E + P SVKDR A+ +I +A + +
Sbjct: 241 KKHVSELIGRTPLVYLNKVTEGCGAYIAVKQEMMQPTASVKDRPALAMIADAEKKNLITP 420
Query: 105 GG-IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
G I+ E ++G+ IS+A + G K + +P ++E+ + A GA + P
Sbjct: 421 GKTILIEPTSGNMGISMAFIGALKGYKVVLTMPSYTSMERRVTMRAFGAELILTDPA--- 591
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
KG + G + PN F
Sbjct: 592 ----------------------------KG-----MGGTVKKAYDLLESTPN----AFML 660
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
QF N AN + H+E TGPEIWE TNG +D FV G+GGTV+GV ++L+ KNPN+K Y +
Sbjct: 661 QQFSNPANTQVHFETTGPEIWEDTNGQVDIFVMGIGSGGTVSGVGQYLKSKNPNVKIYGV 840
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTD 343
+P S + N G P G+G + +D +
Sbjct: 841 EPSESNVLNGGKPG---------------PHHITGNGVGFKPDILDM--DVMDKVLEVSS 969
Query: 344 MEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQ-SIGPGHTIVTILCDSGMRHLS 398
+AV+MAR L +GL +G SS N V A+R+A G IVTI G R+LS
Sbjct: 970 EDAVKMARILALKEGLMVGISSGANTVAALRLASLPENKGKLIVTIHPSFGERYLS 1137
>TC14692 similar to UP|CYSK_CITLA (Q43317) Cysteine synthase
(Beta-pyrazolylalanine synthase) (Beta-PA/CSase)
(L-mimosine synthase) (O-acetylserine sulfhydrylase)
(O-acetylserine (Thiol)-lyase) (CSase) (OAS-TL) ,
partial (62%)
Length = 834
Score = 130 bits (328), Expect = 3e-31
Identities = 82/260 (31%), Positives = 126/260 (47%), Gaps = 4/260 (1%)
Frame = +3
Query: 23 VVAYFFCD-RLCNPSKKKKKKKSK--NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLN 79
V+A FC C+ K ++K+ N + + IGNTPL+ +N + D I K E +
Sbjct: 174 VLAVLFCSLENCSALLKMAEEKTTIANDVTELIGNTPLVYLNRIVDGCVGRIAAKLEMME 353
Query: 80 PGGSVKDRVAVQIIEEALESGQLRRG-GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDD 138
P SVKDR+ +I +A E G ++ G ++ E ++G+T I +A +A A G K + +P
Sbjct: 354 PCSSVKDRIGYSMIADAEEKGLIKPGESVLIEATSGNTGIGLAFMAAAKGYKLIITMPSS 533
Query: 139 AAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQ 198
++E+ I+ A GA + P R A + A + R+ P
Sbjct: 534 MSLERRTILRAFGAELVLTDPA-------------RGMKGAVQKAEEIREKTP------- 653
Query: 199 INGCKSDGHKHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAA 258
+ QFEN AN + HYE TGPEIW+ +NG +DA V+
Sbjct: 654 --------------------NSYILQQFENPANPKVHYETTGPEIWKSSNGKVDALVSGI 773
Query: 259 GTGGTVAGVSKFLQEKNPNI 278
GTGGT+ G K+L+E+NP +
Sbjct: 774 GTGGTITGAGKYLKEQNPGV 833
>BP071870
Length = 430
Score = 67.4 bits (163), Expect = 4e-12
Identities = 51/170 (30%), Positives = 74/170 (43%), Gaps = 1/170 (0%)
Frame = +2
Query: 74 KCEFLNPGGSVKDRVAVQIIEEALESGQLRRG-GIVTEGSAGSTAISIATVAPAYGCKCH 132
K E++ GSVKDR+A+ +IE+A G + RG I+ E ++G+T I +A +A G K
Sbjct: 41 KLEYMQACGSVKDRIALSMIEDAENKGLITRGKTILVEPTSGNTGIGLAFIAAWRGYKVI 220
Query: 133 VVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPK 192
VV+P ++E+ I+ A GA V P K
Sbjct: 221 VVMPSYVSLERKIILRAFGAEVYLTDPA-------------------------------K 307
Query: 193 GTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPE 242
G D + + + G + QFEN AN + HYE TGPE
Sbjct: 308 GVDGV---------FQKAEEIVAETPGSYMLQQFENPANPKIHYETTGPE 430
>TC18952 similar to GB|AAD54324.1|5853119|AF177212 threonine
dehydratase/deaminase {Arabidopsis thaliana;} , partial
(49%)
Length = 951
Score = 46.2 bits (108), Expect = 1e-05
Identities = 59/239 (24%), Positives = 92/239 (37%), Gaps = 1/239 (0%)
Frame = +1
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGI 107
+ D +PL LS+ G I K E L P S K R A ++ + L L +G I
Sbjct: 118 VYDVAIESPLQMATKLSERLGVNIWLKREDLQPVFSFKLRGAYNMMAK-LPKELLEKGVI 294
Query: 108 VTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDH 167
+ SAG+ A +A A C +V+P K + +E +GATV + D
Sbjct: 295 CS--SAGNHAQGVALAAKRLNCSAVIVMPVTTPDIKWKSVEKMGATV-------VLIGDS 447
Query: 168 FVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFE 227
+ EA +A K K +G F F+
Sbjct: 448 Y--------DEAQTYAKKRGKE----------------------------EGRAFIPPFD 519
Query: 228 NLANFRAHYEGT-GPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
+ +GT G EI Q G + A G GG +AG++ +++ +P +K ++P
Sbjct: 520 HPDVIMG--QGTVGMEIVRQMQGPVHAIFVPVGGGGLIAGIAAYVKRVSPQVKIIGVEP 690
>TC10541 similar to UP|CYSL_CAPAN (P31300) Cysteine synthase, chloroplast
precursor (O-acetylserine sulfhydrylase)
(O-acetylserine (Thiol)-lyase) (CSase B) (CS-B) (OAS-TL
B) , partial (25%)
Length = 568
Score = 45.1 bits (105), Expect = 2e-05
Identities = 23/72 (31%), Positives = 40/72 (54%), Gaps = 1/72 (1%)
Frame = +3
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG-G 106
+ IG TP++ +N++ + I K E + P SVKDR+ +I +A + G + G
Sbjct: 351 VTQLIGKTPMVYLNNIVKGSVANIAAKLEIMEPCCSVKDRIGYSMITDAEQRGAITPGKS 530
Query: 107 IVTEGSAGSTAI 118
I+ E ++G+T I
Sbjct: 531 ILVEPTSGNTGI 566
>BP067741
Length = 517
Score = 35.0 bits (79), Expect = 0.023
Identities = 24/81 (29%), Positives = 38/81 (46%), Gaps = 1/81 (1%)
Frame = -2
Query: 319 EGIGINRVTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-Q 377
+GIG + LD + + EAV+ A L +GL +G SSA A+++A +
Sbjct: 513 QGIGAGFIPGTVDVDILDEVVQVSSEEAVDTATLLALKEGLLVGISSAAAAAAAIKLARR 334
Query: 378 SIGPGHTIVTILCDSGMRHLS 398
G I+ + G R+LS
Sbjct: 333 PDNDGKLILVVFPSPGERYLS 271
>TC17110 similar to UP|Q9FS26 (Q9FS26) Plastidic cysteine synthase 1,
partial (28%)
Length = 657
Score = 34.7 bits (78), Expect = 0.031
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 1/81 (1%)
Frame = +1
Query: 319 EGIGINRVTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-Q 377
+GIG V N E +D + EAVE A+ + +GL +G SS A+++ +
Sbjct: 37 QGIGAGFVPGNLDEEVVDEVIAISSDEAVETAKQVALQEGLLVGISSGAAAAAALQVGKR 216
Query: 378 SIGPGHTIVTILCDSGMRHLS 398
G I + G R+LS
Sbjct: 217 PENAGKLIAVVFPSFGERYLS 279
>TC11736
Length = 583
Score = 30.8 bits (68), Expect = 0.44
Identities = 18/53 (33%), Positives = 26/53 (48%), Gaps = 6/53 (11%)
Frame = -2
Query: 32 LCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGCEILGK------CEFL 78
+CN S KKK K + G+ + P++ L + GC I GK C+FL
Sbjct: 501 ICNKSWKKKD*KRQTGLKPSQFMHPILSFQRLENGWGCCIAGKSQSNT*CQFL 343
>TC8274 similar to GB|AAA36350.1|189049|HUMNADH NADH dehydrogenase
(ubiquinone) {Homo sapiens;} , partial (19%)
Length = 1675
Score = 28.5 bits (62), Expect = 2.2
Identities = 16/35 (45%), Positives = 21/35 (59%), Gaps = 3/35 (8%)
Frame = -2
Query: 259 GTGGTVAGVSKFLQEKNPNIKCYLID---PPGSGL 290
G+ GT++G F +K P +CYLID P SGL
Sbjct: 1593 GSPGTLSG---FNHQKKPGTQCYLIDWEFEPDSGL 1498
>TC12009 similar to PIR|T52589|T52589 ribose-phosphate diphosphokinase
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (42%)
Length = 526
Score = 28.1 bits (61), Expect = 2.9
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +3
Query: 37 KKKKKKKSKNGIIDA 51
KKKKKKK KNGII +
Sbjct: 27 KKKKKKKKKNGIISS 71
>AV421085
Length = 348
Score = 27.7 bits (60), Expect = 3.7
Identities = 14/62 (22%), Positives = 29/62 (46%)
Frame = +1
Query: 99 SGQLRRGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVR 158
+G R ++T + +S + KC V P+D +E+ ++ E +G V++
Sbjct: 22 AGVSRSAAVITAYLMKTENLSQEDALESLRQKCEFVCPNDGFLEQLKMFEEMGFKVDQSS 201
Query: 159 PV 160
P+
Sbjct: 202 PI 207
>AW719194
Length = 526
Score = 26.9 bits (58), Expect = 6.4
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = +2
Query: 26 YFFCDRLCNPSKKKKKKKSKN 46
Y+ D+L N KKKKKKK+++
Sbjct: 236 YYIYDKLLNFIKKKKKKKNRH 298
>TC10960 similar to PIR|B96700|B96700 protein F12A21.17 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(11%)
Length = 716
Score = 26.9 bits (58), Expect = 6.4
Identities = 12/32 (37%), Positives = 20/32 (62%)
Frame = +1
Query: 173 RRRASEANEFAFKHRKSQPKGTDSQQINGCKS 204
R+ ASEA E F+ ++ K S++++GC S
Sbjct: 154 RKVASEAGEINFQAQEEGQKSCSSERLSGCCS 249
>BP085925
Length = 334
Score = 26.6 bits (57), Expect = 8.3
Identities = 14/41 (34%), Positives = 21/41 (51%)
Frame = +2
Query: 16 TAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTP 56
T + + + +FF +C SKKKKK +S G + I P
Sbjct: 32 TLVILEWLA*FFF---VCTVSKKKKKLRSHRGDVQGIRQHP 145
>TC19253
Length = 532
Score = 26.6 bits (57), Expect = 8.3
Identities = 15/42 (35%), Positives = 25/42 (58%), Gaps = 2/42 (4%)
Frame = -1
Query: 250 NLDAFVAAAGTGGTVAGVSKFLQEKNPNI-KCY-LIDPPGSG 289
++ A V+ G+GGT++ VSK+ + I C+ L + PG G
Sbjct: 361 SITAMVSRFGSGGTISAVSKYSSARQS*I*ICFRLANAPGKG 236
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,452,392
Number of Sequences: 28460
Number of extensions: 81579
Number of successful extensions: 535
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 501
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 527
length of query: 424
length of database: 4,897,600
effective HSP length: 93
effective length of query: 331
effective length of database: 2,250,820
effective search space: 745021420
effective search space used: 745021420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC122165.6


