

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.2 + phase: 0
(428 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
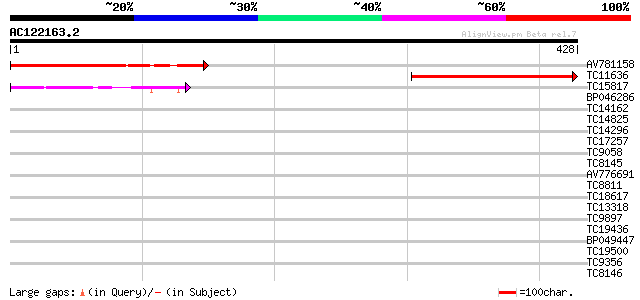
Score E
Sequences producing significant alignments: (bits) Value
AV781158 208 1e-54
TC11636 178 2e-45
TC15817 104 2e-23
BP046286 33 0.12
TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein p... 30 0.76
TC14825 similar to UP|Q8S4W7 (Q8S4W7) GAI-like protein 1, partia... 30 1.00
TC14296 similar to GB|AAM47342.1|21360453|AY113034 AT3g02540/F16... 29 1.3
TC17257 similar to UP|Q89CE1 (Q89CE1) Blr7856 protein, partial (5%) 29 1.3
TC9058 weakly similar to UP|O43809 (O43809) mRNA cleavage factor... 29 1.7
TC8145 weakly similar to UP|Q94A38 (Q94A38) AT5g46250/MPL12_3, p... 29 1.7
AV776691 28 2.2
TC8811 similar to UP|AAP04330 (AAP04330) G-protein-coupled recep... 28 2.2
TC18617 weakly similar to UP|Q9FPQ5 (Q9FPQ5) Gamete-specific hyd... 28 3.8
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 27 4.9
TC9897 similar to UP|ARG1_ARATH (P46637) Arginase , partial (57%) 27 4.9
TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-li... 27 4.9
BP049447 27 6.5
TC19500 27 6.5
TC9356 weakly similar to UP|AGOL_ARATH (Q9SJK3) Argonaute-like p... 27 8.4
TC8146 similar to UP|Q94FP3 (Q94FP3) Succinate dehydrogenase sub... 27 8.4
>AV781158
Length = 511
Score = 208 bits (530), Expect = 1e-54
Identities = 115/152 (75%), Positives = 125/152 (81%), Gaps = 2/152 (1%)
Frame = +3
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MGTRIPSHQLSSGLYVSGRPEQPK + PTMASRSVPYTGGDPKKSGELGKML+IP LDP
Sbjct: 75 MGTRIPSHQLSSGLYVSGRPEQPKTERQPTMASRSVPYTGGDPKKSGELGKMLEIPTLDP 254
Query: 61 -KSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVG 119
K +SSSSQ S+GP RS PNSG G+ +GSG L RKS+GSGPI PTGLITSGP+G
Sbjct: 255 TKPSRASSSSQQSSGPVRSGPNSGHAGRT-SGSGPLPRKSSGSGPIG--PTGLITSGPIG 425
Query: 120 SGPVGASRRSGQLEQ-SGSMGKAVYGSAVTSL 150
G RRSGQLEQ +GS+GKAVYGSAVTSL
Sbjct: 426 GG-----RRSGQLEQPTGSVGKAVYGSAVTSL 506
>TC11636
Length = 655
Score = 178 bits (451), Expect = 2e-45
Identities = 80/125 (64%), Positives = 101/125 (80%)
Frame = +3
Query: 304 DFQTGLRALVKAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLK 363
DFQ+GLRALVK GYG +V P+V + V+DV N+++SP FL WL DRKLS+DDRIMRLK
Sbjct: 3 DFQSGLRALVKTGYGARVTPYVDDSIVIDVNPANKDMSPEFLRWLGDRKLSSDDRIMRLK 182
Query: 364 EGHIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNA 423
EG+IKEGS VSVMGVV+R++NVLMIVPP EP++TGCQW +C+ P +EG+++ CED
Sbjct: 183 EGYIKEGSPVSVMGVVQRNDNVLMIVPPPEPLTTGCQWAKCIFPASLEGIVLRCEDTSKV 362
Query: 424 DVIAV 428
DVI V
Sbjct: 363 DVIPV 377
>TC15817
Length = 773
Score = 104 bits (260), Expect = 2e-23
Identities = 68/144 (47%), Positives = 84/144 (58%), Gaps = 8/144 (5%)
Frame = +3
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+R PSHQLS+GLYVSGRPEQPKER PTM+S ++PYTGGD KKSGELGKM DIP
Sbjct: 291 MGSRFPSHQLSNGLYVSGRPEQPKER-TPTMSSVAMPYTGGDIKKSGELGKMFDIPSDGS 467
Query: 61 KSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPI---ALQPTGLITSGP 117
KS S LS P+R +G+ + ++ SGPI A + +TSG
Sbjct: 468 KSR---KSGPLSNAPSR--------------TGSFAGAASHSGPIMQNAAARSAYVTSGN 596
Query: 118 VGSGPVGAS-----RRSGQLEQSG 136
V +G + S SG L + G
Sbjct: 597 VAAGGMSGSASMKKTNSGPLNKHG 668
>BP046286
Length = 393
Score = 32.7 bits (73), Expect = 0.12
Identities = 25/72 (34%), Positives = 29/72 (39%), Gaps = 3/72 (4%)
Frame = -2
Query: 60 PKSHPSSSSSQLSTGPAR---SRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSG 116
PKS S LS+ P SRP V + G + KS S +P G TS
Sbjct: 281 PKSKSGSWLGTLSSNPKNGISSRPPPRLVSSSKPCPGQSNMKSKSSTGFGSEPVGTTTSA 102
Query: 117 PVGSGPVGASRR 128
VG G G RR
Sbjct: 101 TVGGGDGGNVRR 66
>TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein precursor,
partial (87%)
Length = 1232
Score = 30.0 bits (66), Expect = 0.76
Identities = 21/67 (31%), Positives = 32/67 (47%)
Frame = +2
Query: 60 PKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVG 119
P S PSS+++ +GPA S + T S + S+ P + P+ +T GP
Sbjct: 107 PSS*PSSTTANTPSGPASSPTPA-------TQSSPAAASSSTPSPTSPSPSQTLT-GPAA 262
Query: 120 SGPVGAS 126
SGP A+
Sbjct: 263 SGPAPAA 283
>TC14825 similar to UP|Q8S4W7 (Q8S4W7) GAI-like protein 1, partial (47%)
Length = 1392
Score = 29.6 bits (65), Expect = 1.00
Identities = 27/80 (33%), Positives = 36/80 (44%)
Frame = +3
Query: 62 SHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSG 121
S PSSSS+ S PARSR +S SG SR+ + S L T L++
Sbjct: 363 STPSSSSTSCSRAPARSRRSS-------LSSGKSSRRFSPSWSRKLATTALVS------- 500
Query: 122 PVGASRRSGQLEQSGSMGKA 141
G+ RS + S + KA
Sbjct: 501 *TGSLSRSTTTQPSSTRSKA 560
>TC14296 similar to GB|AAM47342.1|21360453|AY113034 AT3g02540/F16B3_17
{Arabidopsis thaliana;}, partial (76%)
Length = 1567
Score = 29.3 bits (64), Expect = 1.3
Identities = 23/82 (28%), Positives = 33/82 (40%), Gaps = 5/82 (6%)
Frame = +1
Query: 65 SSSSSQLSTGPARSRPNSGQVGKNITGSGTLSR---KSTGSGPIALQPTGL--ITSGPVG 119
S Q P P S Q G + L+ + P+ L P GL + SGP G
Sbjct: 754 SGIPEQAEAPPVTQMPASAQPGNPPAAAPQLANVPSSGPNANPLDLFPQGLPNVGSGPAG 933
Query: 120 SGPVGASRRSGQLEQSGSMGKA 141
+G + R S Q + +M +A
Sbjct: 934 AGSLDFLRNSQQFQALRAMVQA 999
>TC17257 similar to UP|Q89CE1 (Q89CE1) Blr7856 protein, partial (5%)
Length = 614
Score = 29.3 bits (64), Expect = 1.3
Identities = 18/65 (27%), Positives = 28/65 (42%), Gaps = 5/65 (7%)
Frame = +2
Query: 23 PKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP-----KSHPSSSSSQLSTGPAR 77
P+ P ++PY G P+ G G ++ PV+DP +SHP + +
Sbjct: 23 PRGSMPVAGPGDALPYCYGMPQPVGPAGVIMGKPVMDPNMYAQQSHPYMAPPMWPQPQDQ 202
Query: 78 SRPNS 82
RP S
Sbjct: 203QRPPS 217
>TC9058 weakly similar to UP|O43809 (O43809) mRNA cleavage factor I 25 kDa
subunit (Pre-mRNA cleavage factor IM) (25KD), partial
(79%)
Length = 1059
Score = 28.9 bits (63), Expect = 1.7
Identities = 13/43 (30%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Frame = +3
Query: 350 DRKLSTDDRIMRLKEGHIKEGSTVSVMGVV----RRHENVLMI 388
++ S DR+ R+K ++KEG SV G++ H ++L++
Sbjct: 204 EKDTSVADRLARMKVNYMKEGMRTSVEGILLVQEHNHPHILLL 332
>TC8145 weakly similar to UP|Q94A38 (Q94A38) AT5g46250/MPL12_3, partial
(49%)
Length = 1671
Score = 28.9 bits (63), Expect = 1.7
Identities = 13/31 (41%), Positives = 19/31 (60%)
Frame = -2
Query: 81 NSGQVGKNITGSGTLSRKSTGSGPIALQPTG 111
+S ++ I+GS T SR GSGP+ P+G
Sbjct: 239 SSSEIRT*ISGSSTSSRAVAGSGPVTGSPSG 147
>AV776691
Length = 570
Score = 28.5 bits (62), Expect = 2.2
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +2
Query: 52 MLDIPV-LDPKSHPSSSSSQLSTGPARSRPNS--GQVGKNITGSGTLSRKSTGSGPIAL 107
+L+ P+ L P++SS PA P G +G + +G LSR++ GSG +L
Sbjct: 194 LLEGPITLSSAKFPTTSSPPRRLPPATENPPELPGPIGSSSPITGRLSRRTIGSGAYSL 370
>TC8811 similar to UP|AAP04330 (AAP04330) G-protein-coupled receptor GPR34
type 2a (Fragment), partial (6%)
Length = 675
Score = 28.5 bits (62), Expect = 2.2
Identities = 18/57 (31%), Positives = 24/57 (41%)
Frame = +3
Query: 73 TGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRS 129
T P + P+ G TG+GT S +TGS G++ G GP G S
Sbjct: 18 TTPGTTTPSMGTGTGTGTGTGTPSSTTTGSTTPFSSTPGVLGGIGTGMGPSGVGTNS 188
>TC18617 weakly similar to UP|Q9FPQ5 (Q9FPQ5) Gamete-specific
hydroxyproline-rich glycoprotein a2, partial (13%)
Length = 578
Score = 27.7 bits (60), Expect = 3.8
Identities = 29/109 (26%), Positives = 39/109 (35%), Gaps = 3/109 (2%)
Frame = +3
Query: 21 EQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRP 80
+ P PP + S S P P S P P SH S+ + S P P
Sbjct: 129 DSPSPTSPPYLISSSPP----SPSASSSPPPGPTPPPPAPSSHSLSTLAGSSKSPPCLTP 296
Query: 81 ---NSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGAS 126
+ Q + + S +T S P + PTG P+ P GAS
Sbjct: 297 PRHHRHQTPPPQKPTSSASTAATASRPRSASPTGSNRGYPLIPSPHGAS 443
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 27.3 bits (59), Expect = 4.9
Identities = 21/64 (32%), Positives = 28/64 (42%)
Frame = -3
Query: 56 PVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITS 115
P S PSSSSS S+ + S +S + + S + S S+ S TGL
Sbjct: 400 PSTSTSSLPSSSSSSSSSSSSSSPSSSSSSSSSSSSSSSSSSSSSSSSSSLSV*TGL--G 227
Query: 116 GPVG 119
GP G
Sbjct: 226 GPSG 215
>TC9897 similar to UP|ARG1_ARATH (P46637) Arginase , partial (57%)
Length = 696
Score = 27.3 bits (59), Expect = 4.9
Identities = 15/39 (38%), Positives = 18/39 (45%)
Frame = -3
Query: 92 SGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSG 130
SG SR T +GP + T LIT + P RSG
Sbjct: 586 SGCASR*RTSTGPPSFSETALITGNDIEWSPPKTKGRSG 470
>TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-like protein,
partial (19%)
Length = 570
Score = 27.3 bits (59), Expect = 4.9
Identities = 22/87 (25%), Positives = 35/87 (39%)
Frame = +2
Query: 20 PEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSR 79
P P +++SRS DP L + + SSS +T PA SR
Sbjct: 296 PRTPSRASTSSVSSRSATTPSPDPSLISHLSQT-----------SNHSSSTATTSPALSR 442
Query: 80 PNSGQVGKNITGSGTLSRKSTGSGPIA 106
S + + +LSR +T + P++
Sbjct: 443 RQSSP-----STASSLSRSATTTSPVS 508
>BP049447
Length = 478
Score = 26.9 bits (58), Expect = 6.5
Identities = 14/33 (42%), Positives = 16/33 (48%)
Frame = +1
Query: 56 PVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKN 88
P DP SH LST P+ PNS G+N
Sbjct: 313 PQFDPTSHDDFLEQMLSTLPSFWDPNSDLSGEN 411
>TC19500
Length = 448
Score = 26.9 bits (58), Expect = 6.5
Identities = 28/83 (33%), Positives = 36/83 (42%), Gaps = 6/83 (7%)
Frame = -1
Query: 63 HPSSSSSQLSTGPARSR-----PNSGQVGKNITGSGTLSRKSTGSGPIALQPTG-LITSG 116
H SSSSS S + P+S N+ + L ST + P TG LITSG
Sbjct: 442 HHSSSSSHSSAVAVSVKTPSLSPHSLVS*HNVNFNFPLVTPSTSTAPPLAAATGALITSG 263
Query: 117 PVGSGPVGASRRSGQLEQSGSMG 139
P S G ++E GS+G
Sbjct: 262 PDTS---GTGSDESEVEGPGSIG 203
>TC9356 weakly similar to UP|AGOL_ARATH (Q9SJK3) Argonaute-like protein
At2g27880, partial (10%)
Length = 697
Score = 26.6 bits (57), Expect = 8.4
Identities = 19/64 (29%), Positives = 29/64 (44%), Gaps = 2/64 (3%)
Frame = -1
Query: 42 DPKKSGELGKMLDIPVLDPK-SHPSSSSSQLSTGPARSRPNSG-QVGKNITGSGTLSRKS 99
+P +SG L +D S S+S + STG G ++G TG+G +
Sbjct: 370 NPGRSGNLIAFVDDGAAGRNASFCSTSELRASTGTGEGGGGDGAEIGAETTGAGEIGAPM 191
Query: 100 TGSG 103
TG+G
Sbjct: 190 TGAG 179
>TC8146 similar to UP|Q94FP3 (Q94FP3) Succinate dehydrogenase subunit 3
(Fragment), partial (92%)
Length = 1038
Score = 26.6 bits (57), Expect = 8.4
Identities = 26/86 (30%), Positives = 40/86 (46%), Gaps = 4/86 (4%)
Frame = -1
Query: 59 DPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIAL-QPTGLITSGP 117
D H + S+ ST + S+ +S Q ++S +S G+ I + TG ++ G
Sbjct: 348 DSHLHWTIGSTSFSTISSSSQQSSVQ---------SISIESQGAEVIGIFGKTGQVSPGS 196
Query: 118 V---GSGPVGASRRSGQLEQSGSMGK 140
V GSG RRSG +E+ GK
Sbjct: 195 VFTTGSGGCSDLRRSGAVERGEGSGK 118
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,535,771
Number of Sequences: 28460
Number of extensions: 105030
Number of successful extensions: 683
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 669
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 675
length of query: 428
length of database: 4,897,600
effective HSP length: 93
effective length of query: 335
effective length of database: 2,250,820
effective search space: 754024700
effective search space used: 754024700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC122163.2


