

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.9 + phase: 0
(435 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
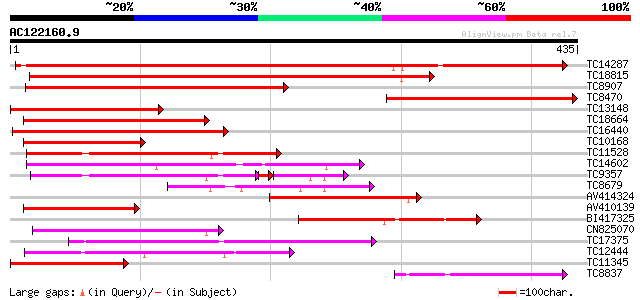
Score E
Sequences producing significant alignments: (bits) Value
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 436 e-123
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 380 e-106
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 280 3e-76
TC8470 homologue to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase,... 215 1e-56
TC13148 similar to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, ... 198 2e-51
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 192 9e-50
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 172 7e-44
TC10168 homologue to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase... 161 2e-40
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 155 2e-38
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 141 2e-34
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 121 5e-33
TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partia... 124 4e-29
AV414324 124 4e-29
AV410139 119 1e-27
BI417325 117 5e-27
CN825070 113 5e-26
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 101 2e-22
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 100 6e-22
TC11345 weakly similar to UP|Q84XC0 (Q84XC0) Calcineurin B-like-... 99 2e-21
TC8837 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein ki... 97 4e-21
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 436 bits (1120), Expect = e-123
Identities = 229/439 (52%), Positives = 303/439 (68%), Gaps = 15/439 (3%)
Frame = +3
Query: 5 ENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIK 64
ENP + ++ KYE+GRVLG G FAKVYH +NL T +NVA+KV+ KEK+ K +++QIK
Sbjct: 402 ENPRN---IIFNKYEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIK 572
Query: 65 REISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQL 124
RE+SVM++V+HP+IV+L EVMA+K KI++ ME V+GGELF K+ KG+L ED AR YFQQL
Sbjct: 573 REVSVMRLVRHPHIVELKEVMATKGKIFLVMEYVKGGELFTKVNKGKLNEDDARKYFQQL 752
Query: 125 ISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYV 184
ISAVDFCHSRGV HRDLKPENLLLDE+ +LKVSDFGL E R DG+L T CGTPAYV
Sbjct: 753 ISAVDFCHSRGVTHRDLKPENLLLDENEDLKVSDFGLSALPEQRRDDGMLVTPCGTPAYV 932
Query: 185 SPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLE 244
+PEV+ KKGYDG+KADIWSCGVILY LL+G+LPFQ +N++ +Y+K ++ +++ P W S +
Sbjct: 933 APEVLKKKGYDGSKADIWSCGVILYALLSGYLPFQGENVMRIYRKAFKAEYEFPEWISPQ 1112
Query: 245 ARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDK----------- 293
A+ LI+ LL +P R SI +I+ WF+ + +A E ED
Sbjct: 1113AKNLISNLLVADPEKRYSIPEIISDPWFQYGFMRPLAFSINESAVGEDSTI*SFWWGGEX 1292
Query: 294 VFEFMEC--EKSSTTMNAFHII-SLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISK 350
V E + + + NAF II SLS GFDL LFE +K R F + + S V++K
Sbjct: 1293VPELVAAMGKPARPFYNAFEIISSLSHGFDLRSLFETRK---RSPSMFISKYSASAVVAK 1463
Query: 351 LEQVAKAVKFDVK-SSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLE 409
LE +AK + F V + VR+QG + GRKGKLA+ +++ V P VVE K GDTLE
Sbjct: 1464LEGMAKKLNFRVTGKKEFTVRMQGMKEGRKGKLAMTMEVFEVAPEVAVVEFSKSAGDTLE 1643
Query: 410 YNQFCSKELRPALKDIFWT 428
Y +FC +++RP+LKDI W+
Sbjct: 1644YVKFCEEQVRPSLKDIVWS 1700
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 380 bits (977), Expect = e-106
Identities = 191/316 (60%), Positives = 238/316 (74%), Gaps = 5/316 (1%)
Frame = +2
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
GKYE+GRVLG GT AKVY AK + +G+ VA+KV+ K ++ K GM++QIKREIS+M++V+H
Sbjct: 32 GKYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVRH 211
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDFCHSRG 135
PNIV L EVMA+K+KI+ ME +RGGELF K+ KG+LK+D+AR YFQQLISAVD+CHSRG
Sbjct: 212 PNIVNLKEVMATKTKIFFIMEYIRGGELFAKVAKGKLKDDLARRYFQQLISAVDYCHSRG 391
Query: 136 VYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYD 195
V HRDLKPENLLLDE+ NLKVSDFGL E LRQDGLLHT CGTPAYV+PEV+ KKGYD
Sbjct: 392 VSHRDLKPENLLLDENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKKGYD 571
Query: 196 GAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDP 255
G K D WSCGVILY LLAG LPFQ +NL+ MY K+ R +F+ PPWFS E+++LI+K+L
Sbjct: 572 GFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLRAEFQFPPWFSPESKKLISKILVA 751
Query: 256 NPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFME----CEKSSTTMNAFH 311
+PN RI+IS IM SWF+K SI + ++ + E ++ +NAF
Sbjct: 752 DPNRRITISSIMRVSWFQKGFSASIPIPDPDESNFNSDLNSSSEQSTKVVAAAKFINAFE 931
Query: 312 II-SLSEGFDLSPLFE 326
I S+S GFDLS FE
Sbjct: 932 FISSMSSGFDLSGFFE 979
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 280 bits (717), Expect = 3e-76
Identities = 131/202 (64%), Positives = 169/202 (82%)
Frame = +3
Query: 13 LLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKM 72
+L KYE+G++LG G FAKVYH +N+ T ++VA+KV+ KE++ K +++QIKRE+SVM++
Sbjct: 327 ILFNKYEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKKERLVKQIKREVSVMRL 506
Query: 73 VKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDFCH 132
V+HP+IV+L EVMA+K+KI++ +E V+GGELF K+ KG++ E AR YFQQLISAVDFCH
Sbjct: 507 VRHPHIVELKEVMATKTKIFMVVEYVKGGELFAKLTKGKMTEVAARKYFQQLISAVDFCH 686
Query: 133 SRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKK 192
SRGV HRDLKPENLLLD++ +LKVSDFGL + E R DG+L T CGTPAYV+PEV+ KK
Sbjct: 687 SRGVTHRDLKPENLLLDDNEDLKVSDFGLSSLPEQRRSDGMLLTPCGTPAYVAPEVLKKK 866
Query: 193 GYDGAKADIWSCGVILYVLLAG 214
GYDG+KADIWSCGVILY LL G
Sbjct: 867 GYDGSKADIWSCGVILYALLCG 932
>TC8470 homologue to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, partial
(29%)
Length = 807
Score = 215 bits (547), Expect = 1e-56
Identities = 111/146 (76%), Positives = 124/146 (84%)
Frame = +1
Query: 290 EEDKVFEFMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVIS 349
E+ K+ E + ++ +TMNAFHIISLSEGFDLSPLFEE+KREE+EEMRFAT S VIS
Sbjct: 1 EKLKLQEQEQVQEQPSTMNAFHIISLSEGFDLSPLFEERKREEKEEMRFATTRPASSVIS 180
Query: 350 KLEQVAKAVKFDVKSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLE 409
+LE+VAKA KFDVK S+ KVRLQGQERGRKGKLAIAAD+YAVTPSFLVVEVKKD GDTLE
Sbjct: 181 RLEEVAKAGKFDVKKSEGKVRLQGQERGRKGKLAIAADMYAVTPSFLVVEVKKDNGDTLE 360
Query: 410 YNQFCSKELRPALKDIFWTSADPPAA 435
YNQFCSKELRPALKDI W + P A
Sbjct: 361 YNQFCSKELRPALKDIVWRTETPTPA 438
>TC13148 similar to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, partial
(24%)
Length = 545
Score = 198 bits (503), Expect = 2e-51
Identities = 100/119 (84%), Positives = 110/119 (92%), Gaps = 1/119 (0%)
Frame = +2
Query: 1 MCEKENPN-SHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGM 59
M +K N +HSA+L GKYELGRVLGHGTFAKVY+A+NL TGKNVAMKV+GKEKVIKVGM
Sbjct: 188 MADKSCSNEAHSAVLQGKYELGRVLGHGTFAKVYNARNLKTGKNVAMKVIGKEKVIKVGM 367
Query: 60 IEQIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVAR 118
+EQIKREISVMKMVKHPNIV LHEVMASKS+IYIAMELVRGGELFNK+ KGRLKED+AR
Sbjct: 368 MEQIKREISVMKMVKHPNIVDLHEVMASKSRIYIAMELVRGGELFNKVAKGRLKEDLAR 544
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 192 bits (488), Expect = 9e-50
Identities = 91/144 (63%), Positives = 118/144 (81%), Gaps = 1/144 (0%)
Frame = +3
Query: 11 SALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVM 70
+ +L GKYE+GR+LG G FAKVYHA+N+ TG++VA+KV+ K+KVI G+ +KRE+S+M
Sbjct: 72 TTVLFGKYEVGRLLGCGAFAKVYHARNIETGQSVAVKVINKKKVIGTGLTGHVKREVSIM 251
Query: 71 KMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKI-VKGRLKEDVARVYFQQLISAVD 129
++HPNIV+LHEV+A+K+KIY ME +GGELF +I KGR ED+AR YFQQLISAV
Sbjct: 252 SRLRHPNIVRLHEVLATKTKIYFVMEFAKGGELFARISTKGRFSEDLARRYFQQLISAVG 431
Query: 130 FCHSRGVYHRDLKPENLLLDEDGN 153
+CHSRGV+HRDLKPENLLLD+ N
Sbjct: 432 YCHSRGVFHRDLKPENLLLDDKVN 503
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 172 bits (437), Expect = 7e-44
Identities = 90/169 (53%), Positives = 120/169 (70%), Gaps = 3/169 (1%)
Frame = +1
Query: 3 EKENPN-SHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIE 61
E+ P+ S ++ GKY+L R LG G FAKVY A +L G VA+K++ K K + M
Sbjct: 43 ERRQPSTSPPRIILGKYQLTRFLGRGNFAKVYQAVSLTDGTTVAVKMIDKSKTVDASMEP 222
Query: 62 QIKREISVMKMVKH-PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARV 119
+I REI M+ ++H PNI+++HEVMA+++KIY+ ++ GGELF+KI + GRL E +AR
Sbjct: 223 RIVREIDAMRRLQHHPNILKIHEVMATRTKIYLIVDYAGGGELFSKISRRGRLPEPLARR 402
Query: 120 YFQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHL 168
YFQQL+SA+ FCH GV HRDLKP+NLLLD +GNLKVSDFGL E L
Sbjct: 403 YFQQLVSALCFCHRNGVAHRDLKPQNLLLDAEGNLKVSDFGLSALPEQL 549
>TC10168 homologue to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, partial
(21%)
Length = 717
Score = 161 bits (407), Expect = 2e-40
Identities = 79/94 (84%), Positives = 89/94 (94%)
Frame = +1
Query: 11 SALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVM 70
S LL GKYELGR+LGHGTFAKVYHA++L +G++VA+KVVGKEKVI+VGM+EQIKREIS M
Sbjct: 436 STLLHGKYELGRLLGHGTFAKVYHARHLESGRSVAIKVVGKEKVIRVGMMEQIKREISAM 615
Query: 71 KMVKHPNIVQLHEVMASKSKIYIAMELVRGGELF 104
MVKHPNIVQLHEVMASK+KIYIAMELVRGGELF
Sbjct: 616 SMVKHPNIVQLHEVMASKTKIYIAMELVRGGELF 717
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 155 bits (391), Expect = 2e-38
Identities = 86/198 (43%), Positives = 124/198 (62%), Gaps = 3/198 (1%)
Frame = +3
Query: 14 LDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMV 73
++ +YE + LG G F A++ NTG+ VA+K + + K I E ++REI + +
Sbjct: 66 MEERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGKKID----ENVQREIINHRSL 233
Query: 74 KHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCH 132
+HPNI++ EV+ + + + I +E GGELF +I GR ED AR +FQQLIS V +CH
Sbjct: 234 RHPNIIRFKEVLLTPTHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCH 413
Query: 133 SRGVYHRDLKPENLLLDEDGN--LKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIA 190
S + HRDLK EN LLD + + LK+ DFG +S+ +T GTPAY++PEV++
Sbjct: 414 SMEICHRDLKLENTLLDGNPSPRLKICDFG---YSKSAILHSQPKSTVGTPAYIAPEVLS 584
Query: 191 KKGYDGAKADIWSCGVIL 208
+K YDG AD+WSCGV L
Sbjct: 585 RKEYDGKVADVWSCGVTL 638
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 141 bits (356), Expect = 2e-34
Identities = 86/268 (32%), Positives = 144/268 (53%), Gaps = 9/268 (3%)
Frame = +2
Query: 14 LDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMV 73
L +Y+L +G G F ++ + + A+K++ K + ++ E M ++
Sbjct: 56 LKSQYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLL 235
Query: 74 K-HPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGR---LKEDVARVYFQQLISAVD 129
HPNI+Q+ +V + + +EL + L ++IV + E A +QL+ AV
Sbjct: 236 SPHPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVA 415
Query: 130 FCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCT-FSEHLRQDGLLHTTCGTPAYVSPEV 188
CH GV HRD+KP+N+L G+LK++DFG F + R G++ GTP YV+PEV
Sbjct: 416 HCHRLGVAHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGRRMSGVV----GTPYYVAPEV 583
Query: 189 IAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWF----SLE 244
+ + Y G K D+WSCGVILY++L+G PF D+ +++ + RG+ + P S
Sbjct: 584 LMGREY-GEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRIFRNVSPA 760
Query: 245 ARRLITKLLDPNPNTRISISKIMDSSWF 272
A+ L+ K++ +P+ RIS + + WF
Sbjct: 761 AKDLLRKMICRDPSNRISAEQALRHPWF 844
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 121 bits (304), Expect(3) = 5e-33
Identities = 74/179 (41%), Positives = 105/179 (58%), Gaps = 3/179 (1%)
Frame = +1
Query: 17 KYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKHP 76
+Y+L R +G G F ++ T + VA+K + + I E +KREI + ++HP
Sbjct: 358 RYDLVRDIGSGNFGVARLMQDKQTKELVAVKYIERGDKID----ENVKREIINHRSLRHP 525
Query: 77 NIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCHSRG 135
NIV+ EV+ + + + I ME GGELF +I GR ED AR +FQQLIS V +CH+
Sbjct: 526 NIVRFKEVILTPTHLAIVMEYASGGELFERICNAGRFTEDEARFFFQQLISGVSYCHAMQ 705
Query: 136 VYHRDLKPENLLLD--EDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKK 192
V HRDLK EN LLD LK+ DFG +S+ +T GTPAY++PEV++K+
Sbjct: 706 VCHRDLKLENTLLDGSPTPRLKICDFG---YSKSSVLHSQPKSTVGTPAYIAPEVLSKQ 873
Score = 35.0 bits (79), Expect(3) = 5e-33
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 6/64 (9%)
Frame = +3
Query: 203 SCGVILYVLLAGFLPFQDDNLVAMYKK----IYRGDFKCPPW--FSLEARRLITKLLDPN 256
SCGV LYV+L G PF+D N ++K + + P + S E R LI+++ +
Sbjct: 906 SCGVTLYVMLVGAYPFEDPNEPKDFRKTIQRVLSVQYSIPDFVQISPECRHLISRIFVFD 1085
Query: 257 PNTR 260
P R
Sbjct: 1086PAER 1097
Score = 21.6 bits (44), Expect(3) = 5e-33
Identities = 7/11 (63%), Positives = 8/11 (72%)
Frame = +2
Query: 192 KGYDGAKADIW 202
K YDG AD+W
Sbjct: 872 KSYDGKIADVW 904
>TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partial (70%)
Length = 972
Score = 124 bits (310), Expect = 4e-29
Identities = 70/171 (40%), Positives = 100/171 (57%), Gaps = 12/171 (7%)
Frame = +2
Query: 122 QQLISAVDFCHSRGVYHRDLKPENLLLDEDG--NLKVSDFGLCTFSEHLRQDGLLHT--- 176
+QLIS V +CH+ + HRDLK EN LLD LK+ DFG S LLH+
Sbjct: 2 EQLISGVHYCHALQICHRDLKLENTLLDGSPAPRLKICDFGYSKSS-------LLHSRPK 160
Query: 177 -TCGTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDD----NLVAMYKKIY 231
T GTPAY++PEV++++ YDG AD+WSCGV LYV+L G PF+D N ++I
Sbjct: 161 STVGTPAYIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDHDDPRNFRKTIQRIM 340
Query: 232 RGDFKCPPW--FSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSI 280
+K P + S + R L++++ NP RIS+ +I + WF K +P+ +
Sbjct: 341 AVQYKIPDYVHISQDCRHLLSRIFVANPLRRISLKEIKNHPWFLKNLPREL 493
>AV414324
Length = 380
Score = 124 bits (310), Expect = 4e-29
Identities = 64/125 (51%), Positives = 83/125 (66%), Gaps = 8/125 (6%)
Frame = +3
Query: 200 DIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPNPNT 259
DIWSCGVILYVLLAG+LPF+D NL+ MY+KI + +FK P WF+ + RRL++K+LDPNP T
Sbjct: 3 DIWSCGVILYVLLAGYLPFRDSNLMEMYRKIGKAEFKFPNWFAPDVRRLLSKILDPNPRT 182
Query: 260 RISISKIMDSSWFKKPVPKSIAMRKKEKE-EEEDKVFEFMECEKSS-------TTMNAFH 311
RIS++KIM+SSWF+K + K ++E D F CE +NAF
Sbjct: 183 RISLAKIMESSWFRKGLQKPAITESGDREIAPLDTDGLFGACENGGPVELSKPCNLNAFD 362
Query: 312 IISLS 316
IIS S
Sbjct: 363 IISYS 377
>AV410139
Length = 435
Score = 119 bits (298), Expect = 1e-27
Identities = 54/89 (60%), Positives = 75/89 (83%)
Frame = +3
Query: 11 SALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVM 70
S LL G++E+G++LGHGTFAKVY+A+N+ G+ VA+KV+ KEK++K G++ IKREIS++
Sbjct: 168 SNLLLGRFEIGKLLGHGTFAKVYYARNIKNGEGVAIKVIDKEKILKGGLVAHIKREISIL 347
Query: 71 KMVKHPNIVQLHEVMASKSKIYIAMELVR 99
+ V+HPNIVQL EVMA+K+KIY ME VR
Sbjct: 348 RRVRHPNIVQLFEVMATKTKIYFVMEYVR 434
>BI417325
Length = 449
Score = 117 bits (292), Expect = 5e-27
Identities = 64/151 (42%), Positives = 92/151 (60%), Gaps = 10/151 (6%)
Frame = +2
Query: 222 NLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIA 281
NL+ +YKKI +F CPPW S AR+LIT++LDPNP TRI+I++I++ WFKK I
Sbjct: 5 NLMELYKKISAAEFTCPPWLSFSARKLITRILDPNPMTRITIAEILEDEWFKKDYKPPIF 184
Query: 282 MRKKE----------KEEEEDKVFEFMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKRE 331
E K+ EE V E + E+ T MNAF +IS+S+G +L LF+ ++
Sbjct: 185 QENGETNLDDVEAVFKDSEEHHVTE--KKEEQPTAMNAFELISMSKGLNLENLFDVEQGF 358
Query: 332 EREEMRFATGGTPSRVISKLEQVAKAVKFDV 362
+R E RF + + +ISK+E+ AK + FDV
Sbjct: 359 KR-ETRFTSKSPANEIISKIEEAAKPLGFDV 448
>CN825070
Length = 634
Score = 113 bits (283), Expect = 5e-26
Identities = 61/152 (40%), Positives = 88/152 (57%), Gaps = 5/152 (3%)
Frame = +3
Query: 18 YELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMK-MVKHP 76
Y GR LG G F Y ++ +TG A K + K K++ + + REI +M + +HP
Sbjct: 129 YTFGRKLGQGQFGTTYLCRHNSTGCTFACKSIPKRKLLCKEDYDDVWREIQIMHHLSEHP 308
Query: 77 NIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSRG 135
++V++H + ++I ME+ GGELF++IV KG+ E A + ++ V+ CHS G
Sbjct: 309 HVVRIHGTYEDAASVHIVMEICEGGELFDRIVQKGQYSEREAAKLIRTIVEVVEACHSLG 488
Query: 136 VYHRDLKPENLLLD---EDGNLKVSDFGLCTF 164
V HRDLKPEN L D ED LK +DFGL F
Sbjct: 489 VMHRDLKPENFLFDTVEEDAKLKTTDFGLSVF 584
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 101 bits (252), Expect = 2e-22
Identities = 64/239 (26%), Positives = 123/239 (50%), Gaps = 3/239 (1%)
Frame = +3
Query: 46 MKVVGKEKVIKVGMIEQIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFN 105
+KV +K K ++Q+ +EI+++ HPNIVQ + + + + +E V GG +
Sbjct: 24 VKVFSDDKTSKE-CLKQLNQEINLLNQFSHPNIVQYYGSELGEESLSVYLEYVSGGSIHK 200
Query: 106 KIVK-GRLKEDVARVYFQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTF 164
+ + G KE V + Y +Q++S + + HSR HRD+K N+L+D +G +K++DFG+
Sbjct: 201 LLQEYGAFKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGM--- 371
Query: 165 SEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPF-QDDNL 223
S+H+ + + G+P +++PEV+ G DI S G + + P+ Q + +
Sbjct: 372 SKHINSAASMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKPPWSQFEGV 551
Query: 224 VAMYKKIYRGDF-KCPPWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIA 281
A++K D + P S +A+ I + L +P R + +++ + + +A
Sbjct: 552 AAIFKIGNSKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIRDQSATKVA 728
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 100 bits (248), Expect = 6e-22
Identities = 64/212 (30%), Positives = 103/212 (48%), Gaps = 5/212 (2%)
Frame = +2
Query: 12 ALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMK 71
+L G Y L +G+G A VY A L + VA+K V ++ +E I+RE M
Sbjct: 71 SLNSGDYNLLEEVGYGASATVYRAVFLPRDEEVAVKCVDLDRCN--ANLEDIRREAQTMS 244
Query: 72 MVKHPNIVQLHEVMASKSKIYIAMELVRGGE---LFNKIVKGRLKEDVARVYFQQLISAV 128
++ H N+V+ H ++++ M + G L +E+ ++ + A+
Sbjct: 245 LIDHRNVVRAHWSFVVDRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSILKETLKAL 424
Query: 129 DFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCT--FSEHLRQDGLLHTTCGTPAYVSP 186
++ H G HRD+K N+LL DG +K++DFG+ F RQ +T GTP +++P
Sbjct: 425 EYLHRHGHIHRDVKAGNILLGSDGQVKLADFGVSASMFDAGERQRN-RNTFVGTPCWMAP 601
Query: 187 EVIAKKGYDGAKADIWSCGVILYVLLAGFLPF 218
EV+ KAD+WS G+ L G PF
Sbjct: 602 EVLQPGTGYNFKADVWSFGITALELAHGHAPF 697
>TC11345 weakly similar to UP|Q84XC0 (Q84XC0) Calcineurin B-like-interacting
protein kinase, partial (15%)
Length = 431
Score = 98.6 bits (244), Expect = 2e-21
Identities = 46/91 (50%), Positives = 68/91 (74%)
Frame = +3
Query: 1 MCEKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMI 60
M E E + L GKYELGRVLG G FAKV++A+N+ TG++VA+K++ K+K+ G+
Sbjct: 159 MPEIEQQPPEATALFGKYELGRVLGCGAFAKVHYARNVQTGQSVAVKIINKKKLAGTGLA 338
Query: 61 EQIKREISVMKMVKHPNIVQLHEVMASKSKI 91
+KREI++M + HP+IV+LHEV+A+K+KI
Sbjct: 339 SNVKREIAIMSRLHHPHIVRLHEVLATKTKI 431
>TC8837 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(15%)
Length = 606
Score = 97.4 bits (241), Expect = 4e-21
Identities = 57/135 (42%), Positives = 80/135 (59%), Gaps = 2/135 (1%)
Frame = +1
Query: 296 EFMECEKSSTTMNAFHIIS-LSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQV 354
+F+E + S NAF IIS LS GFDL LFE +KR F + + S V++KLE V
Sbjct: 31 DFVEAARPS--YNAFEIISSLSHGFDLRNLFETRKRSPST---FISKLSASAVVAKLEAV 195
Query: 355 AKAVKFDVKSS-DTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQF 413
AK + + + VR+QG GRKGK+ + +++ V P VVE K+ GDTLEY +
Sbjct: 196 AKKLNLRITGKKEFMVRMQGVTEGRKGKVGMTVEVFEVAPEVAVVEFSKNAGDTLEYIKL 375
Query: 414 CSKELRPALKDIFWT 428
C ++RP+L DI W+
Sbjct: 376 CEDQVRPSLNDIVWS 420
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,029,010
Number of Sequences: 28460
Number of extensions: 88859
Number of successful extensions: 679
Number of sequences better than 10.0: 206
Number of HSP's better than 10.0 without gapping: 612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 618
length of query: 435
length of database: 4,897,600
effective HSP length: 93
effective length of query: 342
effective length of database: 2,250,820
effective search space: 769780440
effective search space used: 769780440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC122160.9


