

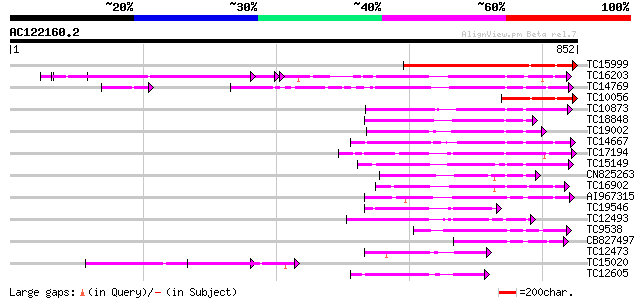
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.2 - phase: 0
(852 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15999 similar to UP|Q9LLT5 (Q9LLT5) Receptor-like protein kina... 475 e-134
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 263 9e-71
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 198 3e-51
TC10056 similar to UP|Q9LLT5 (Q9LLT5) Receptor-like protein kina... 179 1e-45
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 158 3e-39
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 156 1e-38
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 153 1e-37
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 144 8e-35
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 143 1e-34
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 140 8e-34
CN825263 130 9e-31
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 130 1e-30
AI967315 129 2e-30
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 119 2e-27
TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%) 117 1e-26
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 115 4e-26
CB827497 110 9e-25
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 109 2e-24
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 108 4e-24
TC12605 similar to UP|AAR99871 (AAR99871) Strubbelig receptor fa... 103 1e-22
>TC15999 similar to UP|Q9LLT5 (Q9LLT5) Receptor-like protein kinase
(Fragment), complete
Length = 999
Score = 475 bits (1222), Expect = e-134
Identities = 238/263 (90%), Positives = 246/263 (93%), Gaps = 2/263 (0%)
Frame = +1
Query: 592 FLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGV-QSTDDWSTDTWEE 650
FLGRIKHPNLV LTGYC+AGDQRIAIYDYMENGNLQNLLYDLPLGV STDDWSTDTWEE
Sbjct: 1 FLGRIKHPNLVLLTGYCLAGDQRIAIYDYMENGNLQNLLYDLPLGVLHSTDDWSTDTWEE 180
Query: 651 ADN-GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 709
DN GIQN GSEGLLTTW FRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE
Sbjct: 181 PDNNGIQNAGSEGLLTTWSFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 360
Query: 710 PRLSDFGLAKIFGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGK 769
PRLSDFGLAKIFGSGLDEEIARGSPGY PPEF+QP+F++PT KSDVYCFGVVLFELLTGK
Sbjct: 361 PRLSDFGLAKIFGSGLDEEIARGSPGYDPPEFTQPDFDTPTTKSDVYCFGVVLFELLTGK 540
Query: 770 KPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTAD 829
KPV DDY DDKE TLVSWVRGLVRKNQTSRAIDPKI DTG DEQ+EEALK +GYLCTAD
Sbjct: 541 KPVEDDYHDDKE-ETLVSWVRGLVRKNQTSRAIDPKIRDTGPDEQMEEALK-IGYLCTAD 714
Query: 830 LPFKRPTMQQIVGLLKDIEPTTS 852
LPFKRPTMQQIVGLLKDIEP T+
Sbjct: 715 LPFKRPTMQQIVGLLKDIEPATT 783
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 263 bits (672), Expect = 9e-71
Identities = 226/806 (28%), Positives = 374/806 (46%), Gaps = 27/806 (3%)
Frame = +2
Query: 66 EHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSN 124
E++ L ++ LTG IP + +G L KLHSL + N +T T+P + S+ SL SL+LS N
Sbjct: 839 ENLRLLEMANCNLTGEIPPS-LGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSIN 1015
Query: 125 HISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILK 184
++G + + L + +N F +P + L +L+ L++ N F +P +
Sbjct: 1016 DLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGG 1195
Query: 185 CQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISG 243
+ D++ N L+G +P + +L+T + +N G + +S+ + ++
Sbjct: 1196 NGRFLYFDVTKNHLTGLIPPDLCKS-GRLKTFIITDNFFRGPIPKGIGECRSLTKIRVAN 1372
Query: 244 NSFQGSIIE-VFVL-KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLN 301
N G + VF L + +LS N+ G + V + L L LS N +G+I +
Sbjct: 1373 NFLDGPVPPGVFQLPSVTITELSNNRLNGELPSV-ISGESLGTLTLSNNLFTGKIPAAMK 1549
Query: 302 NSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSM 361
N L+ LSL N F + + + L +N+S +L G IP I+H +L A+DLS
Sbjct: 1550 NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR 1729
Query: 362 NHLDGKIPL-LKN-KHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT---LCAS 416
N+L G++P +KN L +++ S N +SGPVP I + + + + S NN T
Sbjct: 1730 NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEI-RFMTSLTTLDLSSNNFTGTVPTGG 1906
Query: 417 EIKPDIMKTSFFGSVN-------SCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALA 469
+ +F G+ N SCP S K R A+V+ ++L A+
Sbjct: 1907 QFLVFNYDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTAR---VRAIVIGIALATAVL 2077
Query: 470 GILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKP 529
+ R++ + +A + + F++
Sbjct: 2078 LVAVTVHVVRKRR--------------------------------LHRAQAWKLTAFQR- 2158
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLV-VGSTLTDEEAAR 588
L I D++ + ++ +G G VYRG +P VA+K LV GS D
Sbjct: 2159 -LEIKAEDVVECLKEEN---IIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRA 2326
Query: 589 ELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTW 648
E+E LG+I+H N++ L GY D + +Y+YM NG+L L+
Sbjct: 2327 EIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLH----------------- 2455
Query: 649 EEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDL 708
G++G W R+KIA+ AR L ++HH CSP IIHR VK++++ LD D
Sbjct: 2456 ----------GAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADF 2605
Query: 709 EPRLSDFGLAK-IFGSGLDEEIA--RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFEL 765
E ++DFGLAK ++ G + ++ GS GY+ PE++ KSDVY FGVVL EL
Sbjct: 2606 EAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYT--LKVDEKSDVYSFGVVLLEL 2779
Query: 766 LTGKKPVGDDYTDDKEATTLVSWV-RGLVRKNQTS------RAIDPKICDTGSDEQIEEA 818
+ G+KPVG + + +V WV + + +Q S +DP++ I
Sbjct: 2780 IIGRKPVG----EFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIH-- 2941
Query: 819 LKEVGYLCTADLPFKRPTMQQIVGLL 844
+ + +C ++ RPTM+++V +L
Sbjct: 2942 MFNIAMMCVKEMGPARPTMREVVHML 3019
Score = 151 bits (381), Expect = 5e-37
Identities = 128/373 (34%), Positives = 187/373 (49%), Gaps = 14/373 (3%)
Frame = +2
Query: 47 YNFSSSV---CSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKI 103
+ FS+S+ CS+ GV CD N VV LN++ + L G +P IG L KL +L +S N +
Sbjct: 266 WKFSTSLSAHCSFSGVTCDQNLR-VVALNVTLVPLFGHLPPE-IGLLEKLENLTISMNNL 439
Query: 104 TT-LPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGL--LENFDLSKNSFSDEIPEALSS 160
T LPSD SLTSLK LN+S N SG NI G+ LE D NSFS +PE +
Sbjct: 440 TDQLPSDLASLTSLKVLNISHNLFSGQFPGNI-TVGMTELEALDAYDNSFSGPLPEEIVK 616
Query: 161 LVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAE 220
L LK L L N F +IP + QSL + L++N L+G +P L+ L+L
Sbjct: 617 LEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLA-KLKTLKELHLGY 793
Query: 221 NNIY-GGV-SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVK 276
+N Y GG+ F ++++ L ++ + G I + KL +L + N G I
Sbjct: 794 SNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPEL 973
Query: 277 YNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLS 336
+ L+ LDLS N L+GEI ++ + NL ++ N+F I L LE L +
Sbjct: 974 SSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVW 1153
Query: 337 KTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI 394
+ + +P + G D++ NHL G IP L K+ L+ + N GP+P I
Sbjct: 1154ENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGI 1333
Query: 395 --LKSLPKMKKYN 405
+SL K++ N
Sbjct: 1334GECRSLTKIRVAN 1372
Score = 135 bits (339), Expect = 4e-32
Identities = 96/310 (30%), Positives = 157/310 (49%), Gaps = 3/310 (0%)
Frame = +2
Query: 63 SNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNL 121
S+ ++ L+LS LTG IP++ KL L ++ NK +LPS L +L++L +
Sbjct: 974 SSMMSLMSLDLSINDLTGEIPES-FSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQV 1150
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSG 181
N+ S L +N+G G FD++KN + IP L LK + N F IP G
Sbjct: 1151 WENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKG 1330
Query: 182 ILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNI 241
I +C+SL I +++N L G +P G P + L+ N + G + + +S+ +L +
Sbjct: 1331 IGECRSLTKIRVANNFLDGPVPPGVFQ-LPSVTITELSNNRLNGELPSVISGESLGTLTL 1507
Query: 242 SGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
S N F G I + L++L L N+F G I + L +++S N L+G I
Sbjct: 1508 SNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTT 1687
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
+ + +L + L+ N + + ++ L+ L LNLS+ + G +PDEI + +L LDL
Sbjct: 1688 ITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDL 1867
Query: 360 SMNHLDGKIP 369
S N+ G +P
Sbjct: 1868 SSNNFTGTVP 1897
Score = 115 bits (288), Expect = 3e-26
Identities = 94/302 (31%), Positives = 146/302 (48%), Gaps = 7/302 (2%)
Frame = +2
Query: 118 SLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRS 177
+LN++ + G L IG LEN +S N+ +D++P L+SL SLKVL + HN+F
Sbjct: 341 ALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQ 520
Query: 178 IPSGI-LKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKS 235
P I + L ++D N SG LP KL+ L+LA N G + ++S +S
Sbjct: 521 FPGNITVGMTELEALDAYDNSFSGPLPEEI-VKLEKLKYLHLAGNYFSGTIPESYSEFQS 697
Query: 236 IVSLNISGNSFQGSIIEVF--VLKLEALDLS-RNQFQGHISQVKYNWSHLVYLDLSENQL 292
+ L ++ NS G + E + L+ L L N ++G I + +L L+++ L
Sbjct: 698 LEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNL 877
Query: 293 SGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLG 352
+GEI +L N L L + N +L G IP E+S +
Sbjct: 878 TGEIPPSLGNLTKLHSLFVQMN------------------------NLTGTIPPELSSMM 985
Query: 353 NLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNN 410
+L +LDLS+N L G+IP K K+L +++F N G +PSFI LP ++ NN
Sbjct: 986 SLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFI-GDLPNLETLQVWENN 1162
Query: 411 LT 412
+
Sbjct: 1163FS 1168
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 198 bits (503), Expect = 3e-51
Identities = 163/522 (31%), Positives = 248/522 (47%), Gaps = 8/522 (1%)
Frame = +3
Query: 333 LNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNKHLQV-IDFSHNNLSGPVP 391
L+LS ++L G IP I+ + NL L++S N DG +P L + +D S+N+L G +P
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSLLISVDLSYNDLMGKLP 1787
Query: 392 SFILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGH 451
I+K LP +K F C + P+ ++NS I + K +
Sbjct: 1788 ESIVK-LPHLKSLYFG------CNEHMSPEDP-----ANMNSSLINTDYGRCKGKESRFG 1931
Query: 452 RGMKLALVLTLSLIFALA-GILFLAFGCRRKNKM--WEVKQGS-YREEQNISGPFSFQTD 507
+ + + + SL+ LA G+LF+ CR + K+ WE G Y E NI FS +
Sbjct: 1932 QVIVIGAITCGSLLITLAFGVLFV---CRYRQKLIPWEGFAGKKYPMETNII--FSLPSK 2096
Query: 508 STTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGN 567
++ SV + F + + AT + TL+ EG FG VYRG L
Sbjct: 2097 DDFFIK------SVSIQAFTLEYIEV-------ATERYK--TLIGEGGFGSVYRGTLNDG 2231
Query: 568 IHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQ 627
VAVKV ST E EL L I+H NLVPL GYC DQ+I +Y +M NG+LQ
Sbjct: 2232 QEVAVKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQ 2411
Query: 628 NLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHG 687
+ LY P ++ + W R IALG AR LA+LH
Sbjct: 2412 DRLYGEP-------------------------AKRKILDWPTRLSIALGAARGLAYLHTF 2516
Query: 688 CSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQP 744
+IHR +K+S++ LD+ + +++DFG +K D ++ RG+ GY+ PE+ +
Sbjct: 2517 PGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKT 2696
Query: 745 EFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDP 804
+ + KSDV+ FGVVL E+++G++P+ + + +LV W +R ++ +DP
Sbjct: 2697 --QQLSEKSDVFSFGVVLLEIVSGREPL--NIKRPRTEWSLVEWATPYIRGSKVDEIVDP 2864
Query: 805 KICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKD 846
I G + + EV C RP+M IV L+D
Sbjct: 2865 GI-KGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELED 2987
Score = 47.4 bits (111), Expect = 1e-05
Identities = 27/78 (34%), Positives = 43/78 (54%)
Frame = +3
Query: 139 LLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQL 198
++ DLS ++ IP +++ + +L+ L + HN F S+PS L L+S+DLS N L
Sbjct: 1596 VITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLS-SLLISVDLSYNDL 1772
Query: 199 SGTLPHGFGDAFPKLRTL 216
G LP P L++L
Sbjct: 1773 MGKLPESI-VKLPHLKSL 1823
Score = 40.4 bits (93), Expect = 0.001
Identities = 26/83 (31%), Positives = 44/83 (52%), Gaps = 1/83 (1%)
Frame = +3
Query: 116 LKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPE-ALSSLVSLKVLKLDHNMF 174
+ L+LSS+++ G + ++I LE ++S NSF +P LSSL L + L +N
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL--LISVDLSYNDL 1772
Query: 175 VRSIPSGILKCQSLVSIDLSSNQ 197
+ +P I+K L S+ N+
Sbjct: 1773 MGKLPESIVKLPHLKSLYFGCNE 1841
Score = 39.7 bits (91), Expect = 0.002
Identities = 26/89 (29%), Positives = 45/89 (50%), Gaps = 2/89 (2%)
Frame = +3
Query: 236 IVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLS 293
I L++S ++ +G I + LE L++S N F G + + S L+ +DLS N L
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLS-SLLISVDLSYNDLM 1775
Query: 294 GEIFQNLNNSMNLKHLSLACNRFSRQKFP 322
G++ +++ +LK L CN + P
Sbjct: 1776 GKLPESIVKLPHLKSLYFGCNEHMSPEDP 1862
Score = 38.1 bits (87), Expect = 0.006
Identities = 31/93 (33%), Positives = 52/93 (55%), Gaps = 4/93 (4%)
Frame = +3
Query: 216 LNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVFVLK--LEALDLSRNQFQGHI 272
L+L+ +N+ G + S+ + + ++ +LNIS NSF GS + F L L ++DLS N G +
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGS-VPSFPLSSLLISVDLSYNDLMGKL 1784
Query: 273 SQVKYNWSHLVYLDLSENQ-LSGEIFQNLNNSM 304
+ HL L N+ +S E N+N+S+
Sbjct: 1785 PESIVKLPHLKSLYFGCNEHMSPEDPANMNSSL 1883
>TC10056 similar to UP|Q9LLT5 (Q9LLT5) Receptor-like protein kinase
(Fragment), partial (48%)
Length = 490
Score = 179 bits (455), Expect = 1e-45
Identities = 89/113 (78%), Positives = 102/113 (89%)
Frame = +2
Query: 740 EFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTS 799
EFSQ EF++P+PKSDVYCFGVVLFE+LTGKKPVGDDY D+K+ TLVSWVRGLVRKN+ S
Sbjct: 2 EFSQQEFDTPSPKSDVYCFGVVLFEILTGKKPVGDDYPDEKKG-TLVSWVRGLVRKNKVS 178
Query: 800 RAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIEPTTS 852
RAID KI DTG +EQ+EEALK +GYLCTADLP KRP+MQQIVGLLKDIEP+ +
Sbjct: 179 RAIDSKIRDTGPEEQMEEALK-IGYLCTADLPSKRPSMQQIVGLLKDIEPSAN 334
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 158 bits (400), Expect = 3e-39
Identities = 107/315 (33%), Positives = 159/315 (49%), Gaps = 4/315 (1%)
Frame = +1
Query: 535 FADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLG 594
F +L AT NF L+ EG FG VY+G L VAVK L +E E+ L
Sbjct: 421 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLS 600
Query: 595 RIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNG 654
+ H NLV L GYC GDQR+ +Y+YM G+L++ L++L + +WST
Sbjct: 601 LLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL-NWST--------- 750
Query: 655 IQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSD 714
R K+A+G AR L +LH PP+I+R +K++++ LD + P+LSD
Sbjct: 751 ---------------RMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSD 885
Query: 715 FGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKP 771
FGLAK+ G + ++ G+ GY PE++ T KSD+Y FGVVL ELLTG++
Sbjct: 886 FGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMS--GKLTLKSDIYSFGVVLLELLTGRRA 1059
Query: 772 VGDDYTDDKEATTLVSWVRGLVR-KNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADL 830
+ D + LVSW R + + +DP + + +A+ + +C +
Sbjct: 1060I--DTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAI-AITAMCLQEQ 1230
Query: 831 PFKRPTMQQIVGLLK 845
P RP + IV L+
Sbjct: 1231PKFRPLITDIVVALE 1275
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 156 bits (395), Expect = 1e-38
Identities = 94/263 (35%), Positives = 139/263 (52%), Gaps = 3/263 (1%)
Frame = +2
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
T+ +L +AT F L EG FG VY G + +AVK L ++ + E A E+E L
Sbjct: 269 TYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVL 448
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLY-DLPLGVQSTDDWSTDTWEEAD 652
GR++H NL+ L GYCV DQR+ +YDYM N +L + L+ + VQ
Sbjct: 449 GRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQ-------------- 586
Query: 653 NGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRL 712
W+ R KIA+G+A + +LHH +P IIHR +KAS+V L+ D EP +
Sbjct: 587 ------------LNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLV 730
Query: 713 SDFGLAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK 770
+DFG AK+ G+ R G+ GY+ PE++ + + DVY FG++L EL+TG+K
Sbjct: 731 ADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAM--WGKVSESCDVYSFGILLLELVTGRK 904
Query: 771 PVGDDYTDDKEATTLVSWVRGLV 793
P+ + T+ W L+
Sbjct: 905 PI--EKLPGGVKRTITEWAEPLI 967
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 153 bits (386), Expect = 1e-37
Identities = 96/272 (35%), Positives = 139/272 (50%), Gaps = 2/272 (0%)
Frame = +1
Query: 537 DLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRI 596
+L SAT+NF+ L EG FG VY G L +AVK L V S D E A E+E L R+
Sbjct: 40 ELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARV 219
Query: 597 KHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQ 656
+H NL+ L GYC G +R+ +YDYM N +L + L+ G S++
Sbjct: 220 RHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLH----GQHSSE--------------- 342
Query: 657 NVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFG 716
L W R IA+G+A + +LHH +P IIHR +KAS+V LD D + R++DFG
Sbjct: 343 ------CLLDWNRRMNIAIGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFG 504
Query: 717 LAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGD 774
AK+ G R G+ GY+ PE++ DV+ FG++L EL +GKKP+
Sbjct: 505 FAKLIPDGATHVTTRVKGTLGYLAPEYAM--LGKANECCDVFSFGILLLELASGKKPL-- 672
Query: 775 DYTDDKEATTLVSWVRGLVRKNQTSRAIDPKI 806
+ ++ W L + + DP++
Sbjct: 673 EKLSSTVKRSINDWALPLACAKKFTEFADPRL 768
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 144 bits (362), Expect = 8e-35
Identities = 112/342 (32%), Positives = 165/342 (47%), Gaps = 4/342 (1%)
Frame = +2
Query: 513 ADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAV 572
A VK T E P L++ +L T NF L+ EG +G VY L VAV
Sbjct: 269 APVKHETQKAPPPIEVPALSLD--ELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAV 442
Query: 573 KVLVVGSTL-TDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLY 631
K L V S T+ E ++ + R+K+ N V L GYCV G+ R+ Y++ G+L ++L+
Sbjct: 443 KKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILH 622
Query: 632 DLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPP 691
GVQ T W IQ V +IA+ AR L +LH P
Sbjct: 623 GRK-GVQGAQPGPTLDW------IQRV-------------RIAVDAARGLEYLHEKVQPA 742
Query: 692 IIHRAVKASSVYLDYDLEPRLSDFGL---AKIFGSGLDEEIARGSPGYVPPEFSQPEFES 748
IIHR +++S+V + D + +++DF L A + L G+ GY PE++
Sbjct: 743 IIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAMT--GQ 916
Query: 749 PTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICD 808
T KSDVY FGVVL ELLTG+KPV D+T + +LV+W + +++ + +DPK+
Sbjct: 917 LTQKSDVYSFGVVLLELLTGRKPV--DHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKG 1090
Query: 809 TGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIEPT 850
+ + + L V LC RP M +V L+ + T
Sbjct: 1091EYPPKGVAK-LAAVAALCVQYEAEFRPNMSIVVKALQPLLKT 1213
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 143 bits (361), Expect = 1e-34
Identities = 113/366 (30%), Positives = 175/366 (46%), Gaps = 8/366 (2%)
Frame = +1
Query: 494 EEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAE 553
++ N S ++T ++ AT + ++ K + ++ +L AT+NF + +
Sbjct: 940 QDGNASSSAEYETSGSSGPGTAS-ATGLTSIMVAKSM-EFSYQELAKATNNFSLDNKIGQ 1113
Query: 554 GKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQ 613
G FG VY L G K+ V ST E EL+ L + H NLV L GYCV G
Sbjct: 1114 GGFGAVYYAELRGKKTAIKKMDVQAST----EFLCELKVLTHVHHLNLVRLIGYCVEGSL 1281
Query: 614 RIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKI 673
+ +Y++++NGNL L+ G E L W R +I
Sbjct: 1282 FL-VYEHIDNGNLGQYLH-------------------------GSGKEPL--PWSSRVQI 1377
Query: 674 ALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIAR-- 731
AL AR L ++H P IHR VK++++ +D +L +++DFGL K+ G R
Sbjct: 1378 ALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLV 1557
Query: 732 GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRG 791
G+ GY+PPE++Q + +PK DVY FGVVLFEL++ K V E+ LV+
Sbjct: 1558 GTFGYMPPEYAQ--YGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEE 1731
Query: 792 LVRKNQTSRA----IDPKICDTGSDEQIEEALK--EVGYLCTADLPFKRPTMQQIVGLLK 845
+ K+ A +DP++ G + I+ LK ++G CT D P RP+M+ +V L
Sbjct: 1732 ALNKSDPCDALRKLVDPRL---GENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALM 1902
Query: 846 DIEPTT 851
+ T
Sbjct: 1903 TLSSLT 1920
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 140 bits (353), Expect = 8e-34
Identities = 101/326 (30%), Positives = 158/326 (47%), Gaps = 1/326 (0%)
Frame = +3
Query: 523 VVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLT 582
+V F DLL A++ +L +G FG Y+ L + VAVK L T++
Sbjct: 225 LVFFGNSARGFDLEDLLRASAE-----VLGKGTFGTAYKAVLETGLVVAVKRLK-DVTIS 386
Query: 583 DEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDD 642
++E ++E +G + H +LVPL Y + D+++ +YDYM G+L LL+
Sbjct: 387 EKEFKDKIETVGAMDHQSLVPLRAYYFSRDEKLLVYDYMPMGSLSALLHG---------- 536
Query: 643 WSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSV 702
N G+ W R IALG AR + +L H P + H +KAS++
Sbjct: 537 --------------NKGAGRTPLNWEIRSGIALGAARGIEYL-HSQGPNVSHGNIKASNI 671
Query: 703 YLDYDLEPRLSDFGLAKIFG-SGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVV 761
L E ++SDFGLA + G S +A GY PE + P + K+DVY FGV+
Sbjct: 672 LLTKSYEAKVSDFGLAHLVGPSSTPNRVA----GYRAPEVTDP--RKVSQKADVYSFGVL 833
Query: 762 LFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKE 821
L ELLTGK P ++E L WV+ LVR+ TS D ++ + E+ L +
Sbjct: 834 LLELLTGKAPT--HALLNEEGVDLPRWVQSLVREEWTSEVFDLELLRYQNVEEEMVQLLQ 1007
Query: 822 VGYLCTADLPFKRPTMQQIVGLLKDI 847
+ C A P +RP++ ++ ++++
Sbjct: 1008LAVDCAAPYPDRRPSIAEVTRSIEEL 1085
>CN825263
Length = 663
Score = 130 bits (327), Expect = 9e-31
Identities = 88/248 (35%), Positives = 121/248 (48%), Gaps = 6/248 (2%)
Frame = +1
Query: 556 FGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRI 615
FG VY+G L VAVK+L E E+E L R+ H NLV L G C+ R
Sbjct: 4 FGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTRC 183
Query: 616 AIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIAL 675
IY+ + NG++++ L+ D W R KIAL
Sbjct: 184 LIYELVPNGSVESHLHGADKETGPLD-------------------------WNARMKIAL 288
Query: 676 GTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDE------EI 729
G AR LA+LH +P +IHR K+S++ L+ D P++SDFGLA+ + LDE
Sbjct: 289 GAARGLAYLHEDSNPCVIHRDFKSSNILLECDFTPKVSDFGLAR---TALDEGNKHISTH 459
Query: 730 ARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWV 789
G+ GY+ PE++ KSDVY +GVVL ELLTG KPV D + LV+W
Sbjct: 460 VMGTFGYLAPEYAMT--GHLLVKSDVYSYGVVLLELLTGTKPV--DLSQPPGQENLVTWA 627
Query: 790 RGLVRKNQ 797
R ++ +
Sbjct: 628 RPILTSKE 651
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 130 bits (326), Expect = 1e-30
Identities = 94/299 (31%), Positives = 142/299 (47%), Gaps = 7/299 (2%)
Frame = +3
Query: 550 LLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDE---EAARELEFLGRIKHPNLVPLTG 606
LL G FG VY G + G VA+K G+ L+++ E E+E L +++H +LV L G
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKR---GNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIG 182
Query: 607 YCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTT 666
YC + I +YD+M G L+ LY ++
Sbjct: 183 YCEENTEMILVYDHMAYGTLREHLYK---------------------------TQKPPLP 281
Query: 667 WRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLD 726
W+ R +I +G AR L +LH G IIHR VK +++ LD ++SDFGL+K G LD
Sbjct: 282 WKQRLEICIGAARGLHYLHTGAKYTIIHRDVKTTNILLDEKWVAKVSDFGLSKT-GPTLD 458
Query: 727 ----EEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEA 782
+ +GS GY+ PE+ + + T KSDVY FGVVLFE+L + + KE
Sbjct: 459 NTHVSTVVKGSFGYLDPEYFRR--QQLTDKSDVYSFGVVLFEILCARPALNPSLA--KEQ 626
Query: 783 TTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIV 841
+L W K + +DP + + E ++ E C +D +RP+M ++
Sbjct: 627 VSLAEWASHCYNKGILDQILDPYLKGKIAPECFKK-FAETAMKCVSDQGIERPSMGDVL 800
>AI967315
Length = 1308
Score = 129 bits (324), Expect = 2e-30
Identities = 94/324 (29%), Positives = 156/324 (48%), Gaps = 9/324 (2%)
Frame = +1
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
++ +L AT+ F ++ +G + VY+G L +AVK L T T + +E EFL
Sbjct: 115 SYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRL----TRTCRDERKEKEFL 282
Query: 594 ------GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDT 647
G + H N++PL G C+ + +++ G++ +L++D +
Sbjct: 283 TEIGTIGHVCHSNVMPLLGCCIDNGLYL-VFELSTVGSVASLIHDEKMAP---------- 429
Query: 648 WEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYD 707
W+ R+KI LGTAR L +LH GC IIHR +KAS++ L D
Sbjct: 430 -----------------LDWKTRYKIVLGTARGLHYLHKGCQRRIIHRDIKASNILLTED 558
Query: 708 LEPRLSDFGLAKIFGS-GLDEEIA--RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFE 764
EP++SDFGLAK S IA G+ G++ PE+ K+DV+ FGV L E
Sbjct: 559 FEPQISDFGLAKWLPSQWTHHSIAPIEGTFGHLAPEYYMHGVVD--EKTDVFAFGVFLLE 732
Query: 765 LLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGY 824
+++G+KPV D +L +W + ++ K + + +DP++ Q +
Sbjct: 733 VISGRKPV------DGSHQSLHTWAKPILSKWEIEKLVDPRLEGCYDVTQFNR-VAFAAS 891
Query: 825 LCTADLPFKRPTMQQIVGLLKDIE 848
LC RPTM +++ ++++ E
Sbjct: 892 LCIRASSTWRPTMSEVLEVMEEGE 963
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 119 bits (298), Expect = 2e-27
Identities = 77/211 (36%), Positives = 110/211 (51%), Gaps = 5/211 (2%)
Frame = +1
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
++ D+ AT NF T+L +G FG VY+ +P VAVKVL S + E E+ L
Sbjct: 202 SYKDIQKATQNFT--TILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQGEHEFQTEVHLL 375
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
GR+ H NLV L G+CV QRI +Y +M NG+L NLLY
Sbjct: 376 GRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLY---------------------- 489
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
G E L +W R +IA+ + + +LH G PP+IHR +K++++ LD + ++
Sbjct: 490 -----GEEKEL-SWDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVA 651
Query: 714 DFGLAK--IF---GSGLDEEIARGSPGYVPP 739
DFGL+K IF SGL +G+ GY+ P
Sbjct: 652 DFGLSKEEIFDGRNSGL-----KGTYGYMDP 729
>TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%)
Length = 837
Score = 117 bits (292), Expect = 1e-26
Identities = 91/286 (31%), Positives = 134/286 (46%), Gaps = 2/286 (0%)
Frame = +3
Query: 507 DSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGT--LLAEGKFGPVYRGFL 564
DS + + + V EK L+ + D + + R + +L +G FG Y+ +
Sbjct: 78 DSESGFVSNSVSAAASVATGEKSLIFVGNVDRVFSLDELLRASAEVLGKGTFGTTYKATM 257
Query: 565 PGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENG 624
VAVK L T T+ E ++E +G++ H NLVPL GY + D+++ +YDYM G
Sbjct: 258 EMGRSVAVKRLK-DVTATEMEFREKIEEVGKLVHENLVPLRGYYFSRDEKLVVYDYMPMG 434
Query: 625 NLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFL 684
+L LL+ A+NG G L W R IALG A +A+L
Sbjct: 435 SLSALLH-------------------ANNG---AGRTPL--NWETRSAIALGAAHGIAYL 542
Query: 685 HHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIARGSPGYVPPEFSQP 744
H P H +K+S++ L EPR+SD GLA + L GY PE +
Sbjct: 543 -HSQGPTSSHGNIKSSNILLTKSFEPRVSDLGLAYL---ALPTSTPNRISGYRAPEVT-- 704
Query: 745 EFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVR 790
+ + K+DVY FG++L ELLTGK P ++E L WV+
Sbjct: 705 DARKVSQKADVYSFGIMLLELLTGKPPTHSSL--NEEGVDLPRWVQ 836
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 115 bits (287), Expect = 4e-26
Identities = 77/243 (31%), Positives = 124/243 (50%), Gaps = 6/243 (2%)
Frame = +2
Query: 608 CVAGD-QRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTT 666
C++ + + +Y+Y+EN +L L+ P + + V + +
Sbjct: 26 CISNEASMLLVYEYLENHSLDKWLHLKP----------------KSSSVSGVVQQYTVLD 157
Query: 667 WRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLD 726
W R KIA+G A+ L+++HH CSPPI+HR VK S++ LD +++DFGLA++ +
Sbjct: 158 WPKRLKIAIGAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGE 337
Query: 727 EEIAR---GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEAT 783
I G+ GY+ PE+ Q S K DVY FGVVL EL TGK+ +Y D +
Sbjct: 338 LNIMSTVIGTFGYIAPEYVQTTRIS--EKVDVYSFGVVLLELTTGKEA---NYGDQHSSL 502
Query: 784 TLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEE--ALKEVGYLCTADLPFKRPTMQQIV 841
+W L+ N D D I+E ++ ++G +CTA LP RP+M++++
Sbjct: 503 AEWAWRHILIGSN----VXDLLXKDVMEASYIDEMCSVFKLGVMCTATLPATRPSMKEVL 670
Query: 842 GLL 844
+L
Sbjct: 671 PIL 679
>CB827497
Length = 550
Score = 110 bits (275), Expect = 9e-25
Identities = 64/176 (36%), Positives = 94/176 (53%), Gaps = 3/176 (1%)
Frame = +2
Query: 667 WRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLD 726
W RH IALGTAR L +LH GC IIHR +KA+++ L D EP++ DFGLAK
Sbjct: 14 WCIRHNIALGTARGLLYLHEGCQRRIIHRDIKAANILLTEDYEPQICDFGLAKWLPENWT 193
Query: 727 EEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEAT 783
G+ GY+ PE+ K+DV+ FGV+L EL++G++ + D
Sbjct: 194 HHTVAKFEGTFGYLAPEYLLHGIVD--EKTDVFAFGVLLLELVSGRRAL------DFSQQ 349
Query: 784 TLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQ 839
+LV W + L++KN+ IDP + D G D + + LC +RP+++Q
Sbjct: 350 SLVLWAKPLLKKNEIMELIDPSLAD-GFDSRQMNLMLLAASLCIQQSSIRRPSIRQ 514
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 109 bits (273), Expect = 2e-24
Identities = 76/201 (37%), Positives = 103/201 (50%), Gaps = 10/201 (4%)
Frame = +3
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFL---------PGN-IHVAVKVLVVGSTLTD 583
+F DL SAT +F L+ EG FG VY+G+L PG+ I VAVK L S
Sbjct: 423 SFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPESMQGF 602
Query: 584 EEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDW 643
E E+ FLGRI HPNLV L GYC ++ + +Y++M G+L+N L+ ++T+
Sbjct: 603 HEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFR-----RNTESL 767
Query: 644 STDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVY 703
S W R KIA+G AR LAFL H +I+R KAS++
Sbjct: 768 S----------------------WNTRLKIAIGAARGLAFL-HSLXKIVIYRDFKASNIL 878
Query: 704 LDYDLEPRLSDFGLAKIFGSG 724
LD + ++S FGL K SG
Sbjct: 879 LDGNYNAKISXFGLTKFGPSG 941
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 108 bits (270), Expect = 4e-24
Identities = 83/260 (31%), Positives = 123/260 (46%), Gaps = 5/260 (1%)
Frame = +1
Query: 115 SLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMF 174
++ + L SGSLT I L DL++NSFS IP +LSSL +LK L L N F
Sbjct: 232 NINQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSF 411
Query: 175 VRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLK 234
SIP I +SL S+D S N LSG+LP+ ++ LR ++L+ N + G +
Sbjct: 412 SGSIPPSITTLKSLYSLDFSHNSLSGSLPNSM-NSLINLRRIDLSFNKLTGSIPKLP--P 582
Query: 235 SIVSLNISGNSFQGSIIEVF---VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQ 291
+++ L + NS G + + + +LE ++LS N G + + L +DLS N
Sbjct: 583 NLLELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNS 762
Query: 292 LSGEIFQN--LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEIS 349
+G NL ++L NR + L L++ SL G IP E
Sbjct: 763 FTGVQISRPARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIPLEYG 942
Query: 350 HLGNLNALDLSMNHLDGKIP 369
+ ++ L L N LDGK P
Sbjct: 943 GIKSMKRLFLDGNFLDGKPP 1002
Score = 80.5 bits (197), Expect = 1e-15
Identities = 60/176 (34%), Positives = 86/176 (48%), Gaps = 8/176 (4%)
Frame = +1
Query: 268 FQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEML 327
+ G ++ + + LV LDL+EN SG I +L++ NLK L+L N FS P I L
Sbjct: 265 YSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSGSIPPSITTL 444
Query: 328 LGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNKHLQVIDFSHNNLS 387
L L+ S SL G +P+ ++ L NL +DLS N L G IP L L+ + N+LS
Sbjct: 445 KSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLPPNLLE-LAVKANSLS 621
Query: 388 GPVPSFILKSLPKMKKYNFSYNNLT--------LCASEIKPDIMKTSFFGSVNSCP 435
GP+ + L +++ S N L L S + D+ SF G S P
Sbjct: 622 GPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNSFTGVQISRP 789
>TC12605 similar to UP|AAR99871 (AAR99871) Strubbelig receptor family 3,
partial (22%)
Length = 680
Score = 103 bits (256), Expect = 1e-22
Identities = 70/210 (33%), Positives = 102/210 (48%), Gaps = 2/210 (0%)
Frame = +3
Query: 513 ADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAV 572
AD +SVP F K T A L T++F + L+ G G V + LP +AV
Sbjct: 66 ADPPTRSSVPPPTFAKCF---TIASLQQYTNSFSQENLIGGGMLGSVXKAELPNGKLLAV 236
Query: 573 KVLVVGSTL--TDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLL 630
L ++ D+E + + RI+H N+V L GYC QR+ IY+Y NG+L + L
Sbjct: 237 XKLDKRASAHQKDDEFLELVNSIDRIRHANIVELIGYCSEHGQRLLIYEYCSNGSLHDAL 416
Query: 631 YDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSP 690
+ S D+ T +W R ++ALG ARAL +LH C P
Sbjct: 417 H-------SDDEIKTKL------------------SWNARIRMALGAARALEYLHELCHP 521
Query: 691 PIIHRAVKASSVYLDYDLEPRLSDFGLAKI 720
P+IHR +K++++ LD DL +SD GL +
Sbjct: 522 PVIHRDLKSANILLDEDLSVHVSDCGLTPL 611
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,698,889
Number of Sequences: 28460
Number of extensions: 211712
Number of successful extensions: 1971
Number of sequences better than 10.0: 362
Number of HSP's better than 10.0 without gapping: 1424
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1677
length of query: 852
length of database: 4,897,600
effective HSP length: 98
effective length of query: 754
effective length of database: 2,108,520
effective search space: 1589824080
effective search space used: 1589824080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122160.2


