

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
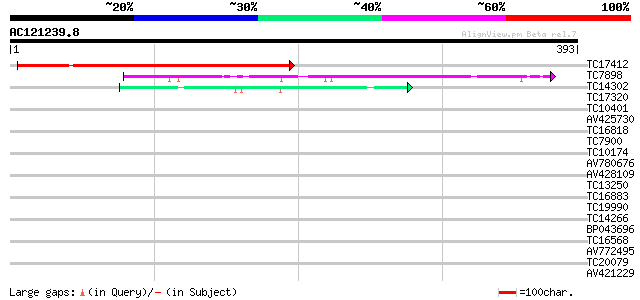
Query= AC121239.8 + phase: 0
(393 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17412 homologue to GB|AAM91372.1|22137054|AY133542 At4g35250/F... 273 4e-74
TC7898 similar to PIR|S17744|S17744 2''-hydroxyisoflavone reduct... 47 7e-06
TC14302 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chl... 41 4e-04
TC17320 similar to UP|IFRI_LUPAL (P52581) Isoflavone reductase h... 38 0.003
TC10401 similar to UP|IFRI_LUPAL (P52581) Isoflavone reductase h... 37 0.004
AV425730 35 0.028
TC16818 similar to UP|Q9ASP7 (Q9ASP7) AT3g46780/T6H20_190, parti... 33 0.082
TC7900 homologue to UP|Q9FUF5 (Q9FUF5) Isoflavone reductase (Fra... 33 0.11
TC10174 32 0.14
AV780676 31 0.31
AV428109 30 0.53
TC13250 weakly similar to UP|Q7NJT5 (Q7NJT5) Glr1747 protein, pa... 30 0.69
TC16883 similar to UP|Q9STI6 (Q9STI6) Nucleotide sugar epimerase... 29 1.5
TC19990 weakly similar to UP|AAQ88099 (AAQ88099) NADPH-dependent... 28 2.6
TC14266 homologue to PIR|C86216|C86216 protein T23G18.6 [importe... 28 3.4
BP043696 28 3.4
TC16568 similar to UP|AAR82759 (AAR82759) RE40656p (Fragment), p... 25 4.0
AV772495 27 4.5
TC20079 UP|Q9FJ12 (Q9FJ12) Genomic DNA, chromosome 5, TAC clone:... 27 4.5
AV421229 27 4.5
>TC17412 homologue to GB|AAM91372.1|22137054|AY133542 At4g35250/F23E12_190
{Arabidopsis thaliana;}, partial (33%)
Length = 578
Score = 273 bits (698), Expect = 4e-74
Identities = 141/193 (73%), Positives = 154/193 (79%), Gaps = 1/193 (0%)
Frame = +2
Query: 6 LMRLPTATPGRVVTRTREAFHALSPSPSPSPPHFVEFCKGGRRGRRIIRVGVKCNSNSER 65
++ LPTA V +R ++P+ P KG RGR +R + +S
Sbjct: 5 VLSLPTAPVSVSVQTSRRYCAVVAPTTVVIPLSLPG--KGRGRGRLRVRCSAEVDSRGGG 178
Query: 66 AV-NLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGAT 124
V N+GPGTPVR T+ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGAT
Sbjct: 179 GVVNMGPGTPVRATNILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGAT 358
Query: 125 VVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFY 184
VVNADL+KPETIPATLVGVHTVIDCATGRPEEPI+TVDWEGKVALIQCAKAMGIQKYVFY
Sbjct: 359 VVNADLTKPETIPATLVGVHTVIDCATGRPEEPIRTVDWEGKVALIQCAKAMGIQKYVFY 538
Query: 185 SIHNCDKHPEVPL 197
SIHNCDKHPEVPL
Sbjct: 539 SIHNCDKHPEVPL 577
>TC7898 similar to PIR|S17744|S17744 2''-hydroxyisoflavone reductase -
alfalfa {Medicago sativa;} , partial (98%)
Length = 1174
Score = 46.6 bits (109), Expect = 7e-06
Identities = 81/336 (24%), Positives = 137/336 (40%), Gaps = 37/336 (11%)
Frame = +1
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVR--------PRPAPA----------DFLRDW 121
ILV+G TG +GR ++ ++ G LVR P+ A D +
Sbjct: 103 ILVIGPTGAIGRHVIWASVKAGNPTYALVRKNSVTIEKPKLITAANPETKEELIDNFKSL 282
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
G ++ D+S E++ + V VI C TGR + +D +V +I K G K
Sbjct: 283 GVILLEGDISDHESLVKAMKQVDIVI-CTTGR----LLILD---QVKIIAAIKEAGNIKR 438
Query: 182 VFYSIH--NCDKHPEV-PLMEIKYCTENFLRDSGLNHIV-------IRLC------GFMQ 225
F S + D+H V P+ E+ F+ +G+ +V LC F++
Sbjct: 439 FFPSEFGLDVDRHEAVDPVREV------FVEKAGIRRVVEAEGIPYTYLCCHAFTGYFLR 600
Query: 226 GLIGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAW- 284
L A +K + D + AY+ D+ T +A + + K + P +
Sbjct: 601 NLAQLDATVPPRDKVIILGDGNVKGAYITEADVGTFTVLAANDPRTLNKAVHIRLPANYL 780
Query: 285 TTQEVITLCERLAGQDANVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFS 344
T E++ L E+ G+ T VP + + + F ++ L S+ L D V+
Sbjct: 781 TANEIMALWEKKIGKTLEKTYVPEEQVLKDIKESGF---PHNYLLALYHSQQLKGDAVYE 951
Query: 345 VPMAETYNL--LGVDTKDIITLEKYLQDYFTNILKK 378
+ A+ L D K T+++YL + F IL+K
Sbjct: 952 IDPAKDAEAHELYPDVK-FTTVDEYL-NQFV*ILRK 1053
>TC14302 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (75%)
Length = 1066
Score = 40.8 bits (94), Expect = 4e-04
Identities = 52/231 (22%), Positives = 90/231 (38%), Gaps = 28/231 (12%)
Frame = +2
Query: 77 PTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGAT-VVNADLSKPET 135
P+++LV GA G G+ + + + Y R LVR + GA V D+ +
Sbjct: 65 PSTVLVTGAGGRTGQIVYKTLKERQYVARGLVRTEESKQKI---GGADDVFIGDIRDAGS 235
Query: 136 IPATLVGVHTVIDCATGRPE-----EPIK---------------TVDWEGKVALIQCAKA 175
I + G+ +I + P+ +P K VDW G+ I AKA
Sbjct: 236 IVPAIQGIDALIILTSATPKMKPGFDPTKGGRPEFYFDDGAYPEQVDWIGQKNQIDAAKA 415
Query: 176 MGIQKYVFYSI-------HNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLI 228
G+++ V H + ++ K E +L DSG+ + +IR G +
Sbjct: 416 AGVKRVVLVGSMGGTNLNHPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRPGGLLDKDG 595
Query: 229 GQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFA 279
G + + ++ + T+ T + D+A + AL E+ K A
Sbjct: 596 GLRELILGKDDELLQTETKT----IPRADVAEVCVQALNYEETQFKAFDLA 736
>TC17320 similar to UP|IFRI_LUPAL (P52581) Isoflavone reductase homolog ,
partial (59%)
Length = 583
Score = 37.7 bits (86), Expect = 0.003
Identities = 24/82 (29%), Positives = 42/82 (50%), Gaps = 7/82 (8%)
Frame = +2
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP-------RPAPADFLRDWGATVVNADL 130
+ +LVVG TG +GR+IV+ +L++G++ L RP + + GA +V A
Sbjct: 38 SKVLVVGGTGYIGRRIVKASLEQGHETYVLQRPELGLQIEKLQMLLSFKKQGAHLVKASF 217
Query: 131 SKPETIPATLVGVHTVIDCATG 152
S +++ + V VI +G
Sbjct: 218 SDHKSLVDAVKKVDVVISAISG 283
>TC10401 similar to UP|IFRI_LUPAL (P52581) Isoflavone reductase homolog ,
partial (49%)
Length = 524
Score = 37.4 bits (85), Expect = 0.004
Identities = 15/33 (45%), Positives = 25/33 (75%)
Frame = +3
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP 110
+ +LVVG TG +GR+IVR ++++G++ L RP
Sbjct: 72 SKVLVVGGTGYIGRRIVRASIEQGHETYVLQRP 170
>AV425730
Length = 426
Score = 34.7 bits (78), Expect = 0.028
Identities = 24/77 (31%), Positives = 37/77 (47%), Gaps = 7/77 (9%)
Frame = +2
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVR----PRPAPADFLRDW---GATVVNADL 130
T ILV+G TG +G+ +V + GY LVR PA A ++ + G +V D+
Sbjct: 71 TKILVIGGTGWIGKFMVEASAKAGYPTFALVRDSTLSSPAKASIIQKFNTLGVNLVLGDI 250
Query: 131 SKPETIPATLVGVHTVI 147
E++ + V VI
Sbjct: 251 HDHESLVKAIKQVDVVI 301
>TC16818 similar to UP|Q9ASP7 (Q9ASP7) AT3g46780/T6H20_190, partial (23%)
Length = 439
Score = 33.1 bits (74), Expect = 0.082
Identities = 16/45 (35%), Positives = 25/45 (55%)
Frame = +2
Query: 61 SNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVR 105
+N+ +LG P+++ V GATG G +I + L EG+ VR
Sbjct: 296 NNNSPGFSLGNARRKDPSTVFVAGATGQAGVRIAQTLLREGFSVR 430
>TC7900 homologue to UP|Q9FUF5 (Q9FUF5) Isoflavone reductase (Fragment),
complete
Length = 509
Score = 32.7 bits (73), Expect = 0.11
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 18/92 (19%)
Frame = +2
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVR--------PRPAPA----------DFLRDW 121
ILV+G TG +GR ++ ++ G LVR P+ A D +
Sbjct: 125 ILVIGPTGAIGRHVIWASVKAGNPTYALVRKNSVTIEKPKLITAANPETKEELIDNFKSL 304
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGR 153
G ++ D+S E++ + V VI C TGR
Sbjct: 305 GVILLEGDISDHESLVKAMKQVDIVI-CTTGR 397
>TC10174
Length = 1063
Score = 32.3 bits (72), Expect = 0.14
Identities = 53/238 (22%), Positives = 92/238 (38%), Gaps = 20/238 (8%)
Frame = +1
Query: 161 VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPLMEIKYCTENFL-----RDSGLNH 215
+D+E + + G +V S C + P + K E L D G ++
Sbjct: 94 IDYEATKNSLVAGRKRGASHFVLLSAI-CVQKPLLEFQRAKLKFEEELMKEAEEDEGFSY 270
Query: 216 IVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAY---MDTQDIAR-LTFIALRNEKI 271
++R F + L GQ V ++++ + R+ + D+A + L +KI
Sbjct: 271 SIVRPTAFFKSLGGQ--VELVKDGKPYVMFGDGRLCACKPISEPDLASFIVDCVLSEDKI 444
Query: 272 NGKLLTFAGP-RAWTTQEVITLCERLAGQDANVTTVPVSV----LRLTRQLTRFFEWTND 326
N K+L GP +A T E + +L G+ N VP+ + + + L + F D
Sbjct: 445 N-KVLPIGGPGKALTPLEQGEMLFKLVGKKPNFLKVPIGIMDFAIGVLDNLVKVFPSLED 621
Query: 327 VAD------RLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILKK 378
A+ A +L D AE G DT L+D+F +L++
Sbjct: 622 AAEFGKIGRYYAVESMLILDPETGEYSAEKTPSYGNDT---------LEDFFARVLRE 768
>AV780676
Length = 526
Score = 31.2 bits (69), Expect = 0.31
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 6/76 (7%)
Frame = +1
Query: 65 RAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRP------RPAPADFL 118
+ V+ P IL++GATG +G+ + + +L G L+RP + A
Sbjct: 58 KMVSTAATPPATAGRILIIGATGFMGQFMTKASLGLGRSTYLLLRPGSLTPSKAAIVKSF 237
Query: 119 RDWGATVVNADLSKPE 134
+D GA V++ ++ E
Sbjct: 238 QDRGAKVIHGVINDKE 285
>AV428109
Length = 168
Score = 30.4 bits (67), Expect = 0.53
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +3
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPA 113
++ V GATG + ++ R L GY VR VR A
Sbjct: 24 TVCVTGATGYVASWLIMRLLQHGYSVRATVRSHHA 128
>TC13250 weakly similar to UP|Q7NJT5 (Q7NJT5) Glr1747 protein, partial (13%)
Length = 575
Score = 30.0 bits (66), Expect = 0.69
Identities = 14/30 (46%), Positives = 18/30 (59%)
Frame = +3
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVR 109
+ V GATG LG ++ L +GY VR L R
Sbjct: 195 VSVTGATGYLGGRLCHALLRQGYSVRILAR 284
>TC16883 similar to UP|Q9STI6 (Q9STI6) Nucleotide sugar epimerase-like
protein, partial (24%)
Length = 602
Score = 28.9 bits (63), Expect = 1.5
Identities = 21/71 (29%), Positives = 32/71 (44%), Gaps = 2/71 (2%)
Frame = +3
Query: 25 FHALSPSPSPSP--PHFVEFCKGGRRGRRIIRVGVKCNSNSERAVNLGPGTPVRPTSILV 82
F LSPSPSPSP P+ G +R+ + + +S+S ++LV
Sbjct: 327 FFILSPSPSPSPTSPNQTRSWGGSDWEKRVTKSARRSSSSS--------------LTVLV 464
Query: 83 VGATGTLGRQI 93
GA G +G +
Sbjct: 465 TGAAGFVGSHV 497
>TC19990 weakly similar to UP|AAQ88099 (AAQ88099) NADPH-dependent cinnamyl
alcohol dehydrogenase, partial (26%)
Length = 421
Score = 28.1 bits (61), Expect = 2.6
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = -1
Query: 35 SPPHFVEFCKGGRRGRRIIRV 55
SPPH E C GG R +I+ +
Sbjct: 142 SPPHLFEICNGGDRSCKILAI 80
>TC14266 homologue to PIR|C86216|C86216 protein T23G18.6 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, complete
Length = 1607
Score = 27.7 bits (60), Expect = 3.4
Identities = 9/29 (31%), Positives = 18/29 (62%)
Frame = +1
Query: 72 GTPVRPTSILVVGATGTLGRQIVRRALDE 100
G P++P +I ++GA G +G + + + E
Sbjct: 58 GNPIKPLTICMIGAGGFIGSHLCEKLMSE 144
>BP043696
Length = 470
Score = 27.7 bits (60), Expect = 3.4
Identities = 17/53 (32%), Positives = 23/53 (43%), Gaps = 16/53 (30%)
Frame = -2
Query: 13 TPGRVVTRTREAFHALSPSPSPSPP----------------HFVEFCKGGRRG 49
+P T R+A A SP P+PSPP + +C+G RRG
Sbjct: 214 SPHASYTPPRKALQASSPPPTPSPPEHRIARSER*RRIRSHRWWRWCRG*RRG 56
>TC16568 similar to UP|AAR82759 (AAR82759) RE40656p (Fragment), partial
(13%)
Length = 421
Score = 25.0 bits (53), Expect(2) = 4.0
Identities = 19/60 (31%), Positives = 25/60 (41%), Gaps = 2/60 (3%)
Frame = -1
Query: 32 PSPSPPHFVEFCKGGRRGRRIIRVGVKCNSNSERAVNLGPG--TPVRPTSILVVGATGTL 89
P P P V FC G G + RV + + + PG P T+ LV+ A G L
Sbjct: 310 PYPPAPP*VVFCSHGLVGTGLARVFLLTAALLSELETISPGLAVPEASTTSLVLPAEGGL 131
Score = 20.8 bits (42), Expect(2) = 4.0
Identities = 7/8 (87%), Positives = 7/8 (87%)
Frame = -1
Query: 30 PSPSPSPP 37
PSP PSPP
Sbjct: 403 PSPPPSPP 380
>AV772495
Length = 572
Score = 27.3 bits (59), Expect = 4.5
Identities = 16/30 (53%), Positives = 18/30 (59%)
Frame = +2
Query: 9 LPTATPGRVVTRTREAFHALSPSPSPSPPH 38
L A P R T T +F SPSPSPSPP+
Sbjct: 425 LSVA*PRRTRTCTYGSF---SPSPSPSPPY 505
>TC20079 UP|Q9FJ12 (Q9FJ12) Genomic DNA, chromosome 5, TAC clone:K21P3
(Extensin like protein), partial (13%)
Length = 530
Score = 27.3 bits (59), Expect = 4.5
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = -3
Query: 21 TREAFHALSPSPSPSPPHFVEFCKGGRRG 49
TR+ AL+PSP+ PP E C G R G
Sbjct: 183 TRQMPRALNPSPAEEPPWTAEGC-GSRTG 100
>AV421229
Length = 262
Score = 27.3 bits (59), Expect = 4.5
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 3/52 (5%)
Frame = +3
Query: 1 MKMATLMRLPTATP--GRVVTRTREAF-HALSPSPSPSPPHFVEFCKGGRRG 49
++ + L++LP P R + RE H+ SPS SP+PP + + G G
Sbjct: 87 LRPSRLIKLPCGGPEANRSIRGPRETPPHSPSPSGSPNPPPILRHHRRGHNG 242
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,345,088
Number of Sequences: 28460
Number of extensions: 92262
Number of successful extensions: 866
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 858
length of query: 393
length of database: 4,897,600
effective HSP length: 92
effective length of query: 301
effective length of database: 2,279,280
effective search space: 686063280
effective search space used: 686063280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC121239.8


