

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
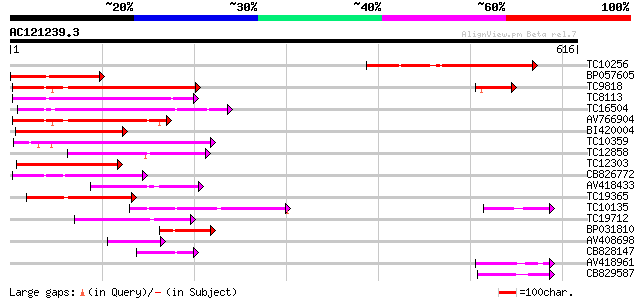
Query= AC121239.3 + phase: 0 /pseudo
(616 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10256 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, part... 249 7e-67
BP057605 143 7e-35
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 136 1e-32
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 132 2e-31
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 114 4e-26
AV766904 110 7e-25
BI420004 99 2e-21
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 97 1e-20
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 91 7e-19
TC12303 similar to GB|CAB10424.1|2245004|ATFCA6 membrane transpo... 89 2e-18
CB826772 87 8e-18
AV418433 75 4e-14
TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter l... 73 1e-13
TC10135 71 6e-13
TC19712 weakly similar to UP|Q9SA38 (Q9SA38) F3O9.19 protein (At... 64 7e-11
BP031810 53 2e-07
AV408698 47 9e-06
CB828147 47 9e-06
AV418961 44 8e-05
CB829587 40 8e-04
>TC10256 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (17%)
Length = 554
Score = 249 bits (637), Expect = 7e-67
Identities = 126/186 (67%), Positives = 154/186 (82%)
Frame = +1
Query: 388 LSNMRQGSLLQGNAGEPVGSTGIGGGWQLAWKWSEQEGPGGKKEGGFKRIYLHQEGGPGS 447
LS+MR+ S L +GEPV STGIGGGWQLAWKWS++ G GK++ GFKRIYLH+E PGS
Sbjct: 16 LSSMRRHSSLMQGSGEPVDSTGIGGGWQLAWKWSDK-GEDGKQKPGFKRIYLHEEAVPGS 192
Query: 448 IRASVVSLPGGDVPTDGDVVQQAAALVSQPALYNKELMHQQPVGPAMIHPSETAAKGPSW 507
R S+VS+PG +G+ VQ AAALVSQPALY+++L+ PVGPAM+HPSETA+K P+W
Sbjct: 193 RRGSIVSIPG-----EGEFVQ-AAALVSQPALYSRDLVGGHPVGPAMVHPSETASKAPTW 354
Query: 508 NDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFL 567
L EPGVKHAL VG+G+Q+LQQFSGINGVLYYTPQIL++AGVG LL+++GLSS SSS L
Sbjct: 355 KVLLEPGVKHALIVGIGIQLLQQFSGINGVLYYTPQILDEAGVGVLLADVGLSSVSSSLL 534
Query: 568 ISAVTT 573
ISA+TT
Sbjct: 535 ISALTT 552
>BP057605
Length = 439
Score = 143 bits (361), Expect = 7e-35
Identities = 72/104 (69%), Positives = 88/104 (84%), Gaps = 1/104 (0%)
Frame = +1
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
M GAV+VA+AA+IGN LQGWDNATIAGS+LYIK++ LQ+ T+EGL+VAMSLIGATV+T
Sbjct: 133 MKGAVLVAIAASIGNFLQGWDNATIAGSVLYIKKDLALQT--TMEGLVVAMSLIGATVIT 306
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLV-MFWSPNVYILLFAR 103
TCSG +SD GRRPMLIISS+LYFL + M WSPNVY+L+ +
Sbjct: 307 TCSGPISDWLGRRPMLIISSVLYFLELVW*MLWSPNVYVLVLRK 438
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 136 bits (342), Expect = 1e-32
Identities = 76/208 (36%), Positives = 130/208 (61%), Gaps = 4/208 (1%)
Frame = +2
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVE---GLIVAMSLIGATVVT 60
A A+ A++ ++L G+D ++G+ +YIKR+ ++ S+ +E G+I SLIG+ +
Sbjct: 236 AFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKV-SDVKIEILLGIINLYSLIGSGL-- 406
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
+G SD GRR ++ + ++F+ +L+M +SPN + L+F R + G+GIG A+ + P+Y
Sbjct: 407 --AGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYWFLMFGRFIAGIGIGYALMIAPVY 580
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMS-LTKAPSWRLMLGVLSIPSLIYFA 179
+E++P RG L + P+ + G+ Y F S L+ WR+MLGV ++PS+I
Sbjct: 581 TAEVSPASSRGFLTSFPEVFINGGILLGYISNFAFSKLSLKVGWRMMLGVGALPSVI-LG 757
Query: 180 LTLLLLPESPRWLVSKGRMLEAKKVLQR 207
+ +L +PESPRWLV +GR+ +A KVL +
Sbjct: 758 VGVLAMPESPRWLVMRGRLGDAIKVLNK 841
Score = 43.5 bits (101), Expect = 1e-04
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 3/47 (6%)
Frame = +2
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV 550
W +LF P ++H + +G+ QQ SGI+ V+ Y+P I E+AG+
Sbjct: 965 WKELFLYPTPAIRHIVIAALGIHFFQQASGIDAVVLYSPTIFEKAGI 1105
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 132 bits (332), Expect = 2e-31
Identities = 73/203 (35%), Positives = 121/203 (58%), Gaps = 1/203 (0%)
Frame = +2
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCS 63
A+ + A++ +++ G+D ++G++L+IK + + T + ++ + I A V +
Sbjct: 116 ALACVIVASMVSIISGYDTGVMSGALLFIKEDIGISD--TQQEVLAGILNICALVGCLAA 289
Query: 64 GALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISE 123
G SD GRR + ++S+L+ + ++ M + PN IL+F R + GLG+G A+T P+Y +E
Sbjct: 290 GKTSDYIGRRYTIFLASILFLVGAVFMGYGPNFAILMFGRCVCGLGVGFALTTAPVYSAE 469
Query: 124 IAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGM-SLTKAPSWRLMLGVLSIPSLIYFALTL 182
++ RG L +LP+ G+F Y + + L WRLMLG+ +IPSL AL +
Sbjct: 470 LSSASTRGFLTSLPEVCIGLGIFIGYISNYFLGKLALTLGWRLMLGLAAIPSL-GLALGI 646
Query: 183 LLLPESPRWLVSKGRMLEAKKVL 205
L +PESPRWLV +GR+ AKKVL
Sbjct: 647 LTMPESPRWLVMQGRLGCAKKVL 715
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 114 bits (286), Expect = 4e-26
Identities = 78/237 (32%), Positives = 135/237 (56%), Gaps = 3/237 (1%)
Frame = +2
Query: 9 VAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTC--SGAL 66
VAA++ L G+ A + G +L+IK + QL S+ V+ ++ L+ + + C +G +
Sbjct: 101 VAASVIFALYGYVMAVMTGVLLFIKEDLQL-SDLQVQ---FSVGLLHMSALPACMTAGRI 268
Query: 67 SDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAP 126
+D GRR +I++++++ + S++M + P+ IL+ + G+G+G ++ + P+Y +EI+P
Sbjct: 269 ADYIGRRYTIILTAIIFLVGSVLMGYGPSYPILMIGLSISGIGLGFSLIVAPVYCAEISP 448
Query: 127 PEIRGSLNTLPQFAGSAGMFFSYCM-VFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLLL 185
P RG L + + + +AG+ Y F L+ WRLML V ++PSL AL +L
Sbjct: 449 PSHRGFLTSFLEVSTNAGIALGYVSNYFFGKLSIRLGWRLMLVVHAVPSLALVAL-MLNW 625
Query: 186 PESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGPDN 242
ESPRWLV +GR+ EA+KVL + ++ A E L + VG D + I+ N
Sbjct: 626 VESPRWLVMQGRVGEARKVLLLVSNTKEEA-EQRLKEIKVAVGIDEKCTQDIVQVPN 793
>AV766904
Length = 611
Score = 110 bits (275), Expect = 7e-25
Identities = 62/178 (34%), Positives = 111/178 (61%), Gaps = 6/178 (3%)
Frame = +3
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVE---GLIVAMSLIGATVVT 60
A+ A+ A++ ++L G+D ++G+++YIKR+ ++ S+ +E G++ S IG+ +
Sbjct: 90 ALACAILASMTSILLGYDIGVMSGAVIYIKRDLKI-SDVQIEILSGIMNIYSPIGSYI-- 260
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
+G SD GRR ++++ L++F+ +++M +SPN L+F R + G+GIG A + P+Y
Sbjct: 261 --AGRTSDWIGRRYTIVLAGLIFFVGAILMGFSPNYAFLMFGRFVAGVGIGFAFLIAPVY 434
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP---SWRLMLGVLSIPSL 175
SEI+P RG L +LP+ +AG+ Y + + +K P WRLMLG+ +IPS+
Sbjct: 435 TSEISPTLSRGFLTSLPEVFLNAGILIGY--ISNYTFSKLPLRLGWRLMLGIGAIPSI 602
>BI420004
Length = 547
Score = 99.0 bits (245), Expect = 2e-21
Identities = 50/123 (40%), Positives = 80/123 (64%), Gaps = 1/123 (0%)
Frame = +1
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQ-LQSEPTVEGLIVAMSLIGATVVTTCSGA 65
V AA IG LL G+D I+G++LYIK +F+ ++ ++ IV+M+L+GA + G
Sbjct: 178 VTAAAGIGGLLFGYDTGVISGALLYIKDDFEEVRRSSLLQETIVSMALVGAIIGAATGGW 357
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
++D FGR+ + + +++ L SLVM +P+ Y ++F RLL GLGIG+A P+YI+E +
Sbjct: 358 INDAFGRKKATLSADVVFTLGSLVMAAAPDAYFVIFGRLLVGLGIGVASVTAPVYIAESS 537
Query: 126 PPE 128
P E
Sbjct: 538 PSE 546
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 96.7 bits (239), Expect = 1e-20
Identities = 73/240 (30%), Positives = 125/240 (51%), Gaps = 21/240 (8%)
Frame = +2
Query: 5 VIVAVAAAIGNLLQGWDNATIAGSIL----YIKREFQLQSEPTV--------------EG 46
+I + AA G L+ G+D ++G + ++ + F + TV +G
Sbjct: 212 IISCILAATGGLMFGYD-VGVSGGVTSMPHFLHKFFPDVYKKTVLEKELDSNYCKYDNQG 388
Query: 47 L-IVAMSLIGATVVTTCSGALSD-LFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARL 104
L + SL A +V+T + + GRR ++I+ + + + + N+ +L+ R+
Sbjct: 389 LQLFTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFFIAGVVFNVTAQNLAMLIVGRI 568
Query: 105 LDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SW 163
L G G+G A VPL++SEIAP IRG+LN L Q + G+ F+ + +G + K W
Sbjct: 569 LLGCGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITIGILFANLINYGTNKIKGGWGW 748
Query: 164 RLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVE 223
RL LG+ IP+++ + LL+ ++P L+ +GR E K VL+++RG +V E LVE
Sbjct: 749 RLSLGLAGIPAVL-LTVGALLVVDTPNSLIERGRTEEGKAVLKKIRGTDNVEPEFLELVE 925
Score = 35.4 bits (80), Expect = 0.027
Identities = 12/31 (38%), Positives = 20/31 (63%)
Frame = +2
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQILEQAG 549
+ + + LQI QQF+GIN +++Y P + G
Sbjct: 995 IIISIALQIFQQFTGINAIMFYAPVLFNTLG 1087
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 90.5 bits (223), Expect = 7e-19
Identities = 53/160 (33%), Positives = 91/160 (56%), Gaps = 4/160 (2%)
Frame = +3
Query: 63 SGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYIS 122
+ ++ + G R +I+ L + +L + +++L+ RLL G GIG A VP+Y+S
Sbjct: 12 ASTITRIMGMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIYLS 191
Query: 123 EIAPPEIRGSLNTLPQFAGSAGMF----FSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYF 178
E+AP + RG LN Q + + G+F F+Y + + WRL LG+ ++P++I F
Sbjct: 192 EMAPYKYRGGLNMCFQLSITIGIFVANLFNY---YFAKILNGQGWRLSLGLGAVPAVI-F 359
Query: 179 ALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEM 218
+ L LP+SP LV++GR A++ L ++RG D+ E+
Sbjct: 360 VVGSLCLPDSPSSLVARGRHEAARQELVKIRGTTDIEAEL 479
Score = 32.3 bits (72), Expect = 0.23
Identities = 14/45 (31%), Positives = 21/45 (46%)
Frame = +3
Query: 507 WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVG 551
W L E + L V + QQF+G+N + +Y P + G G
Sbjct: 528 WKTLLERKYRPQLVFAVCIPFFQQFTGLNVITFYAPILFRTIGFG 662
>TC12303 similar to GB|CAB10424.1|2245004|ATFCA6 membrane transporter like
protein {Arabidopsis thaliana;}, partial (25%)
Length = 545
Score = 89.0 bits (219), Expect = 2e-18
Identities = 47/116 (40%), Positives = 72/116 (61%), Gaps = 1/116 (0%)
Frame = +3
Query: 8 AVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-VEGLIVAMSLIGATVVTTCSGAL 66
A +A IG LL G+D I+G++LYIK +F+ T ++ IV+ ++ GA + G +
Sbjct: 198 AFSAGIGGLLFGYDTGVISGALLYIKDDFKAVDRKTWLQEAIVSTAIAGAIIGAAVGGWI 377
Query: 67 SDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYIS 122
+D FGRR +II+ L+ + S++M +PN ILLF R+ G G+G+A PLYIS
Sbjct: 378 NDRFGRRVAIIIADTLFLIGSVIMAAAPNPAILLFGRVFVGFGVGMASMASPLYIS 545
>CB826772
Length = 490
Score = 87.0 bits (214), Expect = 8e-18
Identities = 44/146 (30%), Positives = 86/146 (58%)
Frame = +1
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCS 63
A + +AA+I + + G++ + G++L+IK + Q+ S+ ++ L+ + G + +
Sbjct: 55 ACVSVMAASILSAIYGYEVGVMTGALLFIKEDLQI-SDLQLQFLVGIFHMCGLPA-SMAA 228
Query: 64 GALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISE 123
G +SD GRR +I++S+ + S++ +SP+ IL+ + G+G G + + PLY +E
Sbjct: 229 GRISDYIGRRYTIILASIAFLSGSILTGYSPSYLILMIGNFISGIGAGFTLIVAPLYCAE 408
Query: 124 IAPPEIRGSLNTLPQFAGSAGMFFSY 149
I+PP RGSL +L +F+ + G+ Y
Sbjct: 409 ISPPSHRGSLTSLTEFSINIGILLGY 486
>AV418433
Length = 401
Score = 74.7 bits (182), Expect = 4e-14
Identities = 48/123 (39%), Positives = 70/123 (56%)
Frame = +3
Query: 88 LVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFF 147
LV+++S +L R+ G G+G+ +VP++I+EIAP E+RG+L TL QF + M
Sbjct: 15 LVIYFSQGPVLLDIGRVATGYGMGVFSYVVPVFIAEIAPKELRGALTTLNQFMLVSAMSV 194
Query: 148 SYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQR 207
S+ V G L SWR + IP+ + L L +PESPRWL +G + + LQ
Sbjct: 195 SF--VIGNVL----SWRALALTGLIPTAV-LLLGLFFIPESPRWLAKRGCREDFEAALQI 353
Query: 208 LRG 210
LRG
Sbjct: 354 LRG 362
>TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (24%)
Length = 441
Score = 73.2 bits (178), Expect = 1e-13
Identities = 42/120 (35%), Positives = 72/120 (60%), Gaps = 1/120 (0%)
Frame = +2
Query: 19 GWDNATIAGSILYIKREFQLQS-EPTVEGLIVAMSLIGATVVTTCSGALSDLFGRRPMLI 77
G+ +AG++L+IK E + + + G I+ + + A +V +G SD GRR +I
Sbjct: 89 GYVTGVMAGALLFIKEELGINDMQVQLLGGILNVCALPACMV---AGRTSDYVGRRYTII 259
Query: 78 ISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLP 137
+S+++ L S++M + P+ IL+ R GLG+GLA+ + P+Y +EI+ P RG L +LP
Sbjct: 260 LSAVILLLGSILMGYGPSFPILMIGRCTAGLGVGLALIIAPVYSTEISLPSNRGFLTSLP 439
>TC10135
Length = 1501
Score = 70.9 bits (172), Expect = 6e-13
Identities = 56/180 (31%), Positives = 90/180 (49%), Gaps = 5/180 (2%)
Frame = +2
Query: 131 GSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLLLPESP 189
G+L +L F + G F SY + ++ T AP +WR MLGV + P++I L +L LPESP
Sbjct: 2 GALVSLNSFLITGGQFLSY--LINLAFTNAPGTWRWMLGVAAAPAIIQIVL-MLSLPESP 172
Query: 190 RWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGPDNELADEED 249
RWL KGR E+K +L+++ +DV E+ L E + ++ IEE + + +
Sbjct: 173 RWLYRKGREEESKSILKKIYAPEDVDAEIEALKESV----ESEIEESKTSKISMMTLLKT 340
Query: 250 PSTGKDQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVT----LFGSV 305
+ + G + Q +V S +V G +N + L+ L+T FGS+
Sbjct: 341 TTVRRGLYAGMGLQFFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLITSGLNAFGSI 520
Score = 48.1 bits (113), Expect = 4e-06
Identities = 27/78 (34%), Positives = 45/78 (57%)
Frame = +2
Query: 515 VKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTL 574
V+ L+ G+GLQ QQF GIN V+YY+P I++ A G +S ++ L+S +T+
Sbjct: 347 VRRGLYAGMGLQFFQQFVGINTVMYYSPTIVQLA---------GFASNRTALLLSLITSG 499
Query: 575 LMLPCIAVAMRLMDISGR 592
L +++ +D +GR
Sbjct: 500 LNAFGSILSIYFIDKTGR 553
>TC19712 weakly similar to UP|Q9SA38 (Q9SA38) F3O9.19 protein
(At1g16390/F3O9_19), partial (35%)
Length = 655
Score = 63.9 bits (154), Expect = 7e-11
Identities = 41/131 (31%), Positives = 65/131 (49%)
Frame = +2
Query: 71 GRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIR 130
GR+ ML S L+ LSSL+ +S N++I + L G G T + SE+ + R
Sbjct: 20 GRKNMLFFSCLMMSLSSLLASFSSNIWIYSSLKFLSGFGRATIGTSSLVLASELVGKKRR 199
Query: 131 GSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLLLPESPR 190
++ F + G M + + + SWR + SIP+++Y L L + ESPR
Sbjct: 200 AQISVCGFFLFTIGFLSLPAMAY---INRNSSWRNLYLWTSIPTILYCILVKLFVQESPR 370
Query: 191 WLVSKGRMLEA 201
WL+ +G+ EA
Sbjct: 371 WLLVRGKKEEA 403
>BP031810
Length = 393
Score = 52.8 bits (125), Expect = 2e-07
Identities = 27/61 (44%), Positives = 42/61 (68%)
Frame = +3
Query: 163 WRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLV 222
WRL LG+ ++P+ + F + LL PE+P LV +GR+ EA++VL+R+RG +V E +V
Sbjct: 69 WRLSLGLATVPATVMF-IGGLLCPETPNSLVEQGRLEEARQVLERVRGTPNVDAEYEDIV 245
Query: 223 E 223
E
Sbjct: 246 E 248
>AV408698
Length = 199
Score = 47.0 bits (110), Expect = 9e-06
Identities = 24/63 (38%), Positives = 35/63 (55%)
Frame = +1
Query: 107 GLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLM 166
G GIG VPLY+SE+AP + RG++N L QF AG+ + + + WR+
Sbjct: 10 GGGIGFGNQAVPLYLSEMAPAKTRGAVNQLFQFTTCAGILIANLVNYFTEKIHPHGWRIS 189
Query: 167 LGV 169
LG+
Sbjct: 190 LGL 198
>CB828147
Length = 570
Score = 47.0 bits (110), Expect = 9e-06
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Frame = +1
Query: 138 QFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKG 196
+F+ + G+ Y F + WR+ML V + PS+ L +L L ESPRWL+ +G
Sbjct: 1 EFSINIGILLGYMSTFFFEKLRLNLGWRMMLAVPAAPSIALIFL-MLKLGESPRWLIMQG 177
Query: 197 RMLEAKKVL 205
R+ +AKKVL
Sbjct: 178 RVGDAKKVL 204
Score = 36.2 bits (82), Expect = 0.016
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Frame = +1
Query: 497 PSETAAKGPSWNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV 550
P +T G + +LF P V+ + VGL + GI +L Y+P+I E+ G+
Sbjct: 307 PKKTRHGGGALKELFCKPTPLVRRIMVAAVGLHVFMHLGGIGVILLYSPRIFEKTGI 477
>AV418961
Length = 394
Score = 43.9 bits (102), Expect = 8e-05
Identities = 29/86 (33%), Positives = 39/86 (44%)
Frame = +3
Query: 507 WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSF 566
W DLF + VG L + QQ +GIN V+YY+ + AG+ S ++S
Sbjct: 36 WFDLFSTRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIA--------SDVAASA 191
Query: 567 LISAVTTLLMLPCIAVAMRLMDISGR 592
L+ A AVA LMD GR
Sbjct: 192 LVGASNVF----GTAVASSLMDRQGR 257
>CB829587
Length = 548
Score = 40.4 bits (93), Expect = 8e-04
Identities = 27/84 (32%), Positives = 38/84 (45%)
Frame = +2
Query: 509 DLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLI 568
DLF ++ +GVGL + QQ GING+ +YT + AGV SS
Sbjct: 107 DLFHTRYVRSVVIGVGLMVCQQSVGINGIGFYTAETFVAAGV------------SSGKAG 250
Query: 569 SAVTTLLMLPCIAVAMRLMDISGR 592
+ + +P + LMD SGR
Sbjct: 251 TIAYACIQVPFTILGAILMDKSGR 322
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,794,190
Number of Sequences: 28460
Number of extensions: 154580
Number of successful extensions: 945
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 913
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 922
length of query: 616
length of database: 4,897,600
effective HSP length: 96
effective length of query: 520
effective length of database: 2,165,440
effective search space: 1126028800
effective search space used: 1126028800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC121239.3


