

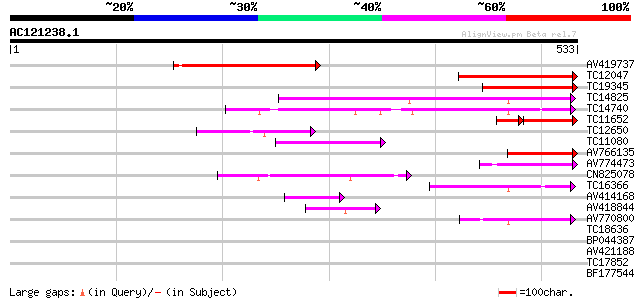
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.1 + phase: 0
(533 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV419737 234 2e-62
TC12047 similar to UP|O81316 (O81316) F6N15.20 protein (SCARECRO... 208 2e-54
TC19345 similar to UP|O81316 (O81316) F6N15.20 protein (SCARECRO... 150 6e-37
TC14825 similar to UP|Q8S4W7 (Q8S4W7) GAI-like protein 1, partia... 122 1e-28
TC14740 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial... 104 4e-23
TC11652 similar to UP|O81316 (O81316) F6N15.20 protein (SCARECRO... 91 7e-22
TC12650 similar to UP|O23210 (O23210) SCARECROW-like protein, pa... 80 1e-15
TC11080 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin r... 77 7e-15
AV766135 70 8e-13
AV774473 69 1e-12
CN825078 68 3e-12
TC16366 similar to PIR|E96542|E96542 scarecrow-like protein [imp... 52 2e-07
AV414168 47 8e-06
AV418844 44 7e-05
AV770800 43 1e-04
TC18636 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial... 38 0.004
BP044387 35 0.030
AV421188 33 0.088
TC17852 similar to UP|Q8W4I8 (Q8W4I8) SCARECROW gene regulator, ... 33 0.15
BF177544 33 0.15
>AV419737
Length = 410
Score = 234 bits (598), Expect = 2e-62
Identities = 117/138 (84%), Positives = 125/138 (89%)
Frame = +3
Query: 155 QKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRA 214
+KV S TGDD + QLQQSIFDQL+KTAELIEAGNPV AQGILARLNHQLSP G PF RA
Sbjct: 3 EKVDS--TGDDVNNQLQQSIFDQLYKTAELIEAGNPVHAQGILARLNHQLSPNGKPFHRA 176
Query: 215 SFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALE 274
+FYMKEALQ MLHSNG+N FSPISFIFKIGAYKSFSEISPVLQFANFTCNQ+LIEA+E
Sbjct: 177 AFYMKEALQSMLHSNGHNFLTFSPISFIFKIGAYKSFSEISPVLQFANFTCNQALIEAVE 356
Query: 275 RFDRIHVIDFDIGFGVQW 292
RFDRIH+IDFDIGFGVQW
Sbjct: 357 RFDRIHIIDFDIGFGVQW 410
>TC12047 similar to UP|O81316 (O81316) F6N15.20 protein (SCARECROW-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (16%)
Length = 617
Score = 208 bits (530), Expect = 2e-54
Identities = 98/111 (88%), Positives = 105/111 (94%)
Frame = +3
Query: 423 VLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGF 482
VLQCYSALLESLDAVNVN DVLQKIERH++QP I KIVL HH+ ++KL PWRN+FLQSGF
Sbjct: 3 VLQCYSALLESLDAVNVNXDVLQKIERHFVQPAIKKIVLGHHHSQEKLTPWRNLFLQSGF 182
Query: 483 SPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
SPF+FSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC
Sbjct: 183 SPFTFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 335
>TC19345 similar to UP|O81316 (O81316) F6N15.20 protein (SCARECROW-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (12%)
Length = 520
Score = 150 bits (378), Expect = 6e-37
Identities = 67/89 (75%), Positives = 78/89 (87%)
Frame = +1
Query: 445 QKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPV 504
QKIERH+I P + K + H++ +KLPPWR+MFLQ GFSPF+FSNFTE+QAECLVQRAPV
Sbjct: 1 QKIERHFILPDMKKTIFGHNHSHEKLPPWRSMFLQYGFSPFTFSNFTESQAECLVQRAPV 180
Query: 505 RGFQVERKPSSLVLCWQRKELISVSTWRC 533
RGFQ+ERK SS VLCWQ+KELISVSTWRC
Sbjct: 181 RGFQLERKHSSFVLCWQQKELISVSTWRC 267
>TC14825 similar to UP|Q8S4W7 (Q8S4W7) GAI-like protein 1, partial (47%)
Length = 1392
Score = 122 bits (306), Expect = 1e-28
Identities = 79/287 (27%), Positives = 132/287 (45%), Gaps = 7/287 (2%)
Frame = +2
Query: 253 EISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKIT 312
E P L+FA+FT NQ+++EA + DR+HVIDF I G+QW + MQ + LR G P+ ++T
Sbjct: 2 ETCPYLKFAHFTANQAILEAFQGKDRVHVIDFAINQGMQWPALMQALALRPGGPPAFRLT 181
Query: 313 AVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIG 372
+ P+ N L L+Q A+ +++ FE+ SL + E++
Sbjct: 182 GIGPPAPDNSDHLQQVGWKLAQLAETIHVQFEYRGFVANSLADLDASMLDLRPPEEESVA 361
Query: 373 VN--FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSAL 430
VN F + ++ P ++Q++P+I+ +++ L YSAL
Sbjct: 362 VNSVFELHKLLARPGALEKVFSVIRQVQPEILTVVEQEAGHNGASFLDRFTESLHYYSAL 541
Query: 431 LESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQR----DKLPPWRNMFLQSGFSPFS 486
+SL+ + + + Y+ I +V + R + L WR F +G SP
Sbjct: 542 FDSLEGSSAVESQDKAMTEIYLGKQICNVVACEGSDRIERHETLTQWRTRFNSAGLSPVH 721
Query: 487 FSNFTEAQAECLVQR-APVRGFQVERKPSSLVLCWQRKELISVSTWR 532
+ QA L+ A G++VE L L W + LI+ S W+
Sbjct: 722 LGSNAFKQASMLLALFAGGDGYRVEENNGCLTLGWHTRPLIATSAWQ 862
>TC14740 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial (57%)
Length = 1406
Score = 104 bits (259), Expect = 4e-23
Identities = 90/356 (25%), Positives = 153/356 (42%), Gaps = 27/356 (7%)
Frame = +1
Query: 204 LSPIGNPFQRASFYMKEALQLMLHSNGNNL---------TAFSPISFIFKIGAYKSFSEI 254
+S G+P QR Y+ E L+ L +G+ + T +S++ + +I
Sbjct: 10 VSVAGDPSQRLGAYLLEGLRARLELSGSLIYKALKCEQPTGKELMSYMHIL------YQI 171
Query: 255 SPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAV 314
P +FA + N ++ EA+ RIH+IDF I G QW +Q + R G P ++IT V
Sbjct: 172 CPYWKFAYISANAAIAEAMANESRIHIIDFQIAQGTQWHLLIQALAHRPGGPPFIRITGV 351
Query: 315 VSPSSCNEI--ELNFTQENLSQYAKDLNILFEFNV-----LNIESLNLPSCPLPGHFFDS 367
S + L E LS +A+ + FEF+ +E NL P
Sbjct: 352 DDSQSFHARGGGLEIVGERLSDFARSYGVPFEFHSAAMSGCEVERENLGVRP-------- 507
Query: 368 NEAIGVNFPV------SSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVV 421
EA+ VNFP +S + L +K L PK+V ++ + P +
Sbjct: 508 GEALAVNFPYVLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLAEQESNTNTSPFFNRFM 687
Query: 422 HVLQCYSALLESLDAVNVNLDVLQ-KIERHYIQPTINKIVLSHHNQR----DKLPPWRNM 476
L+ Y+A+ ES+D D + E+H + I +V +R + W+
Sbjct: 688 ETLEYYTAMFESIDVACPRDDKKRISAEQHCVARDIVNMVACEGAERVERHELFGKWKLR 867
Query: 477 FLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
F +GF + S+ + L+ R +++E++ +L L W ++ L++ S WR
Sbjct: 868 FSMAGFKQWPLSSSVMTATQNLL-RDFNHNYRLEQRDGALCLGWMKRALVTSSAWR 1032
>TC11652 similar to UP|O81316 (O81316) F6N15.20 protein (SCARECROW-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (10%)
Length = 447
Score = 90.9 bits (224), Expect(2) = 7e-22
Identities = 40/50 (80%), Positives = 45/50 (90%)
Frame = +3
Query: 484 PFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
P FSNFTE+QAECLVQR PVRGF VE++ SSLVLCWQRK+LIS+STWRC
Sbjct: 84 PCHFSNFTESQAECLVQRTPVRGFHVEKRQSSLVLCWQRKDLISISTWRC 233
Score = 30.0 bits (66), Expect(2) = 7e-22
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +2
Query: 458 KIVLSHHNQRDKLPPWRNMFLQSGFS 483
K+VL ++ PW+NM L SGFS
Sbjct: 5 KLVLGRQRLQETTLPWKNMLLSSGFS 82
>TC12650 similar to UP|O23210 (O23210) SCARECROW-like protein, partial (23%)
Length = 414
Score = 79.7 bits (195), Expect = 1e-15
Identities = 48/118 (40%), Positives = 71/118 (59%), Gaps = 6/118 (5%)
Frame = +2
Query: 176 DQLFKTAELIEAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEALQLMLHSNGNNLT 234
++L + A+ ++ AQ IL RLN +L SP G P RA+F+ K+ALQ +L +G+N T
Sbjct: 62 EELIRAADCFDSNQLQLAQAILERLNQRLRSPSGKPLHRAAFHFKDALQSLL--SGSNRT 235
Query: 235 AFSP-----ISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIG 287
P + I +K+FS ISP+ F+ FT NQ+L+EAL +HV+DF+IG
Sbjct: 236 NAPPRLSSMAEIVQSIRIFKAFSGISPIPMFSIFTTNQALLEALHGSLFMHVVDFEIG 409
>TC11080 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin response
modulator, partial (33%)
Length = 556
Score = 77.0 bits (188), Expect = 7e-15
Identities = 38/103 (36%), Positives = 57/103 (54%)
Frame = +2
Query: 251 FSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLK 310
F E P L+FA+FT NQ+++EA R+HV+DF + G+QW + MQ + LR G P+ +
Sbjct: 185 FYESCPYLKFAHFTANQAILEAFATAGRVHVVDFGLKQGMQWPALMQALALRPGGPPTFR 364
Query: 311 ITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESL 353
+T + P N L L+Q A+ + + FEF SL
Sbjct: 365 LTGIGPPQPDNSDALQQVGWKLAQLAQTIGVQFEFRGFVCNSL 493
>AV766135
Length = 300
Score = 70.1 bits (170), Expect = 8e-13
Identities = 26/65 (40%), Positives = 41/65 (63%)
Frame = -1
Query: 469 KLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISV 528
++PPWR F +G P S F + QAECL+ ++ +RGF V ++ + LVL W + +++
Sbjct: 291 RMPPWREAFYCAGMRPVQLSQFADFQAECLLAKSQIRGFHVAKRQAELVLFWHERAMVAT 112
Query: 529 STWRC 533
S WRC
Sbjct: 111 SAWRC 97
>AV774473
Length = 449
Score = 69.3 bits (168), Expect = 1e-12
Identities = 32/92 (34%), Positives = 50/92 (53%)
Frame = -3
Query: 442 DVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQR 501
D ++IE ++P KI + + PPWR F +G P S F + QAECL+ +
Sbjct: 444 DWARRIEMLLLRP---KIFAAVEAAGRRSPPWREAFYGAGMRPVQLSQFADFQAECLLAK 274
Query: 502 APVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
+RGF V ++ + LVL W + +++ S WRC
Sbjct: 273 VQIRGFHVAKRQAELVLFWHERAMLATSAWRC 178
>CN825078
Length = 650
Score = 68.2 bits (165), Expect = 3e-12
Identities = 52/188 (27%), Positives = 83/188 (43%), Gaps = 6/188 (3%)
Frame = +3
Query: 196 ILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNN----LTAFSPISFIFKIGAYKSF 251
++ + +S G P QR YM E L ++GN+ L P + +
Sbjct: 39 LIEKARSAVSINGEPIQRLGAYMVEGLVARKEASGNSIYHALNCREPEGEEL-LSYMQLL 215
Query: 252 SEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKI 311
EI P L+F N ++ EA DRIH+IDF I G QW + +Q + R G P ++I
Sbjct: 216 FEICPYLKFGYMAANGAIAEACRNEDRIHIIDFQIAQGSQWMTLLQALAARPGGAPHVRI 395
Query: 312 TAVVSPSS--CNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNE 369
T + P S L+ + L+ ++ I EF+ + + ++ L E
Sbjct: 396 TGIDDPVSKYARGDGLDIVGKRLALMSEKFGIPVEFHGVPVFGPDVTRDMLD---IRPGE 566
Query: 370 AIGVNFPV 377
A+ VNFP+
Sbjct: 567 ALAVNFPL 590
>TC16366 similar to PIR|E96542|E96542 scarecrow-like protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(27%)
Length = 711
Score = 52.0 bits (123), Expect = 2e-07
Identities = 34/144 (23%), Positives = 68/144 (46%), Gaps = 6/144 (4%)
Frame = +3
Query: 395 KQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQ-KIERHYIQ 453
K L PK+V +++ + + P V + Y A+ ES+D V + +E+H +
Sbjct: 6 KCLSPKVVTLVEQEFNTNNAPFLQRFVETMNYYQAVFESIDVVLPREHKERINVEQHCLA 185
Query: 454 PTINKIVLSHHNQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRG-FQ 508
+ +V +R + L WR F +GF+P+ +++ + + L++ RG +
Sbjct: 186 REVVNLVACEGAERVERHELLNKWRMRFASAGFTPYPLNSYINSSIKDLLE--SYRGHYT 359
Query: 509 VERKPSSLVLCWQRKELISVSTWR 532
+E + +L L W + L++ WR
Sbjct: 360 LEERDGALFLGWMNQVLVASCAWR 431
>AV414168
Length = 208
Score = 47.0 bits (110), Expect = 8e-06
Identities = 20/56 (35%), Positives = 32/56 (56%)
Frame = +1
Query: 259 QFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAV 314
+F + N ++ EA++ +H+IDF I G QW S +Q + R G P ++IT V
Sbjct: 4 KFGYMSANGAIAEAMKEEREVHIIDFQISQGTQWVSLIQALAHRPGGPPKVRITGV 171
>AV418844
Length = 284
Score = 43.9 bits (102), Expect = 7e-05
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 2/72 (2%)
Frame = +1
Query: 279 IHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAV--VSPSSCNEIELNFTQENLSQYA 336
+H+IDF I +G QW ++ + R G P L++T + P + T L Y
Sbjct: 16 VHIIDFGICYGFQWPCLIKSLSERHGGPPRLRVTGIELPRPGFRPAERVEETGRRLENYC 195
Query: 337 KDLNILFEFNVL 348
K + FE+N L
Sbjct: 196 KKFKVPFEYNCL 231
>AV770800
Length = 514
Score = 43.1 bits (100), Expect = 1e-04
Identities = 30/113 (26%), Positives = 46/113 (40%), Gaps = 4/113 (3%)
Frame = -1
Query: 424 LQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQR----DKLPPWRNMFLQ 479
L YS + +SL+ V D + + Y+Q I +V R + L WR +
Sbjct: 472 LHYYSTVFDSLEGCPVEPD--KALAEMYLQREICNVVCCEGTARVERHEPLAKWRERLGK 299
Query: 480 SGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
+GF P + QA L G+ VE K L L W + ++ S W+
Sbjct: 298 AGFRPVHLGSDAFTQASMLFTLFSSEGYCVEEKEGCLTLGWHSRPPLAASAWQ 140
>TC18636 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial (10%)
Length = 813
Score = 38.1 bits (87), Expect = 0.004
Identities = 17/61 (27%), Positives = 29/61 (46%)
Frame = +1
Query: 473 WRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
WR+ F GF P S+ A ++ +++E K +L L W+ + + + S WR
Sbjct: 109 WRSRFSMXGFVPCPLSSSVTASVRNILNEFN-ENYRLEHKDVALYLTWKNRAMCTASAWR 285
Query: 533 C 533
C
Sbjct: 286 C 288
>BP044387
Length = 378
Score = 35.0 bits (79), Expect = 0.030
Identities = 17/65 (26%), Positives = 28/65 (42%)
Frame = -2
Query: 468 DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELIS 527
+KL W F +GF S F + L+ G+++ + + CW + L S
Sbjct: 299 EKLGKWVPRFDSTGFGKVPLSYFGMLKTRRLLPSYGCEGYRMRGENGCVANCWPDRSLFS 120
Query: 528 VSTWR 532
+S WR
Sbjct: 119 ISAWR 105
>AV421188
Length = 199
Score = 33.5 bits (75), Expect = 0.088
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Frame = +2
Query: 291 QWSSFMQEIVLRSNGKPSLKITAV--VSPSSCNEIELNFTQENLSQYAKDLNILFEFNVL 348
QW ++ ++ R G P LKIT + P ++ T L+ Y K N+ FE+N +
Sbjct: 5 QWPILIKFLLERDGGPPKLKITGIEFPLPGFRPTERIDETGHRLTNYCKRFNVPFEYNAI 184
>TC17852 similar to UP|Q8W4I8 (Q8W4I8) SCARECROW gene regulator, partial
(26%)
Length = 456
Score = 32.7 bits (73), Expect = 0.15
Identities = 33/142 (23%), Positives = 58/142 (40%), Gaps = 13/142 (9%)
Frame = +2
Query: 371 IGVNFPVSSFI---SNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCY 427
+ VNF + + P+ AL K L+P+IV + V + L+ +
Sbjct: 2 LAVNFMLQLYNLLDETPTAVETALRLAKSLKPRIVTLGEYEASLTRVGFVSRFKAALKHF 181
Query: 428 SALLESLDAVNVNLDVLQKIE----------RHYIQPTINKIVLSHHNQRDKLPPWRNMF 477
SAL ESL+ N+ D ++ + I P + V +++ WR +
Sbjct: 182 SALFESLEP-NLPSDSPERFQVESLLLGRRIAGVIGPELPGCVRERMEDKEQ---WRVLM 349
Query: 478 LQSGFSPFSFSNFTEAQAECLV 499
GF S S++ +QA+ L+
Sbjct: 350 ESCGFESVSLSHYAISQAKILL 415
>BF177544
Length = 318
Score = 32.7 bits (73), Expect = 0.15
Identities = 20/75 (26%), Positives = 33/75 (43%)
Frame = +3
Query: 361 PGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNV 420
PG N A ++ +S + L +K L PK+V ++++ + P
Sbjct: 48 PGEALVVNFAFQLHHMPDESVSTVNERDQLLRLVKSLNPKLVTVVEQDVNTNTAPFLQRF 227
Query: 421 VHVLQCYSALLESLD 435
V + YSA+ ESLD
Sbjct: 228 VEAYKYYSAVFESLD 272
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,397,378
Number of Sequences: 28460
Number of extensions: 132617
Number of successful extensions: 1016
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 949
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 986
length of query: 533
length of database: 4,897,600
effective HSP length: 95
effective length of query: 438
effective length of database: 2,193,900
effective search space: 960928200
effective search space used: 960928200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC121238.1


