

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.2 - phase: 0
(463 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
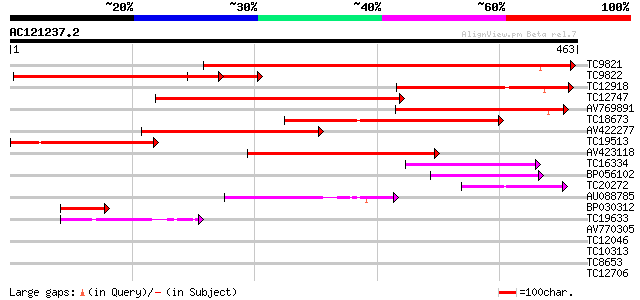
Score E
Sequences producing significant alignments: (bits) Value
TC9821 similar to UP|Q84LI7 (Q84LI7) Polygalacturonase-like prot... 448 e-127
TC9822 similar to UP|Q84LI7 (Q84LI7) Polygalacturonase-like prot... 235 1e-62
TC12918 221 2e-58
TC12747 weakly similar to UP|Q8LAH2 (Q8LAH2) Polygalacturonase-l... 213 5e-56
AV769891 187 4e-48
TC18673 similar to UP|Q9LWY0 (Q9LWY0) ESTs AU029388(E30287), par... 184 3e-47
AV422277 177 3e-45
TC19513 similar to UP|Q84LI7 (Q84LI7) Polygalacturonase-like pro... 149 1e-36
AV423118 132 9e-32
TC16334 101 3e-22
BP056102 87 6e-18
TC20272 weakly similar to UP|Q9LRY8 (Q9LRY8) Polygalacturonase-l... 65 3e-11
AU088785 59 2e-09
BP030312 51 5e-07
TC19633 similar to UP|Q7NET9 (Q7NET9) Glr3789 protein, partial (4%) 48 4e-06
AV770305 32 0.29
TC12046 31 0.49
TC10313 similar to UP|Q8LB12 (Q8LB12) Seh1-like protein, partial... 28 3.2
TC8653 similar to UP|AAS46232 (AAS46232) Methionine sulfoxide re... 28 4.2
TC12706 UP|Q7X9C0 (Q7X9C0) NIN-like protein 2, complete 28 4.2
>TC9821 similar to UP|Q84LI7 (Q84LI7) Polygalacturonase-like protein,
partial (61%)
Length = 1080
Score = 448 bits (1153), Expect = e-127
Identities = 201/308 (65%), Positives = 254/308 (82%), Gaps = 4/308 (1%)
Frame = +3
Query: 159 GSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTIL 218
G+ WW KFH KQL TRPY+IEIMYSD +QISNLTL+NSP+W +HP+Y SN+I+ G+TIL
Sbjct: 15 GALWWKKFHNKQLQYTRPYLIEIMYSDNVQISNLTLVNSPSWNIHPVYCSNVIVQGITIL 194
Query: 219 APVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTC 278
APV SPNTDGINPDSCTN RIED +IVSGDDC+A+KSGWDEYGI GMP++ ++IRRLTC
Sbjct: 195 APVTSPNTDGINPDSCTNTRIEDCYIVSGDDCVAVKSGWDEYGISYGMPTKHLVIRRLTC 374
Query: 279 ISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTM 338
ISP SA++ALGSEMSGGI+DVR ED+ AIN+ES +RIK+AVGRG +V+DI+V+ M + TM
Sbjct: 375 ISPTSAVIALGSEMSGGIEDVRAEDILAINSESGVRIKTAVGRGGYVRDIYVRRMTMKTM 554
Query: 339 KYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSN 398
K+VFWMTG YG H DN +DPNA+P I INYRD+ A+NVT A +LEGIS PFTGIC+S+
Sbjct: 555 KWVFWMTGDYGSHADNNYDPNAIPVIENINYRDMVAENVTTAPRLEGISGAPFTGICISH 734
Query: 399 VTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELL----QEKEIECPFPTDKLAIENVQFK 454
VT+E++ KK+PW CTD+SG++S V P+PCELL +EK C FP D LA+E+VQ +
Sbjct: 735 VTLELAQKAKKVPWTCTDVSGISSGVTPEPCELLPGQAEEKFGACSFPEDHLALEDVQVQ 914
Query: 455 TCNFQSSV 462
TC ++ ++
Sbjct: 915 TCTYRRNL 938
>TC9822 similar to UP|Q84LI7 (Q84LI7) Polygalacturonase-like protein,
partial (40%)
Length = 915
Score = 235 bits (599), Expect = 1e-62
Identities = 112/171 (65%), Positives = 135/171 (78%)
Frame = +1
Query: 4 VISVVLILGSLAECRVPSSIKLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFNAAI 63
++ ++++L SL + L NF+Y A+NCR +S +TDFG VGDG TLNT+AF AI
Sbjct: 49 LVFMMVVLPSLHVVESVRTPVLENFEYCAVNCRAYSPSVTDFGGVGDGTTLNTQAFRKAI 228
Query: 64 TNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPSY 123
+LSQY+++GG+QL VPPG+WLTGSFNLTSHFTLF+ KDAVILGSQDE+EWP + LPSY
Sbjct: 229 EHLSQYSSNGGSQLYVPPGRWLTGSFNLTSHFTLFIHKDAVILGSQDENEWPVIDPLPSY 408
Query: 124 GRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLT 174
GRGRD GRFSSLI GT+LTDVIITGDNGT+DGQG WW KFH KQL T
Sbjct: 409 GRGRDTQGGRFSSLICGTHLTDVIITGDNGTLDGQGDLWWKKFHNKQLQYT 561
Score = 62.4 bits (150), Expect = 2e-10
Identities = 32/61 (52%), Positives = 39/61 (63%)
Frame = +2
Query: 146 VIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPI 205
VI G N TI SY + RPY+IEIMYSD +QISNLTL+NSP+W +HP+
Sbjct: 515 VICGGKNSTIS---SY----------NIPRPYLIEIMYSDNVQISNLTLVNSPSWNIHPV 655
Query: 206 Y 206
Y
Sbjct: 656 Y 658
>TC12918
Length = 545
Score = 221 bits (564), Expect = 2e-58
Identities = 106/146 (72%), Positives = 121/146 (82%), Gaps = 2/146 (1%)
Frame = +1
Query: 317 SAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKN 376
+AVGRGA+VK+IFVKGM+L TMKYVFWMTGSYG HPD GFDP ALP I+GINYRDV AKN
Sbjct: 1 TAVGRGAYVKNIFVKGMNLFTMKYVFWMTGSYGSHPDTGFDPKALPTITGINYRDVIAKN 180
Query: 377 VTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELLQEK- 435
VT KLEGI+NDPFTGIC+SN IE KKL WNCTD+ GVTSNV P+PC LLQEK
Sbjct: 181 VTYPAKLEGIANDPFTGICISNANIEKVG--KKLAWNCTDVHGVTSNVSPEPCALLQEKP 354
Query: 436 -EIECPFPTDKLAIENVQFKTCNFQS 460
+ +CPFP+DKL IE+VQ KTC+F+S
Sbjct: 355 GKFDCPFPSDKLLIESVQLKTCSFKS 432
>TC12747 weakly similar to UP|Q8LAH2 (Q8LAH2) Polygalacturonase-like
protein, partial (43%)
Length = 624
Score = 213 bits (542), Expect = 5e-56
Identities = 103/203 (50%), Positives = 139/203 (67%)
Frame = +2
Query: 120 LPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMI 179
LPSYGRG D P GR+ SLI+G NLTDV+ITG +GTIDGQGS WWD L +RP++I
Sbjct: 11 LPSYGRGIDIPGGRYRSLIYGNNLTDVVITGYDGTIDGQGSVWWDLISTGDLNYSRPHII 190
Query: 180 EIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRI 239
E++ SD I ISNLT++NSP+W +HP+Y + I+ +T+ AP +SP+T GI PDS + I
Sbjct: 191 ELIGSDNITISNLTILNSPSWGIHPVYCRYVQIHNITVHAPPESPHTSGIVPDSSEYICI 370
Query: 240 EDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDV 299
+ + I +G D IA+KSGWDEYG+ G P+ + I + S A +A GSEMSGGI ++
Sbjct: 371 DKSNISTGHDAIALKSGWDEYGVAYGKPTLNVHISSVYLQSSSGAGLAFGSEMSGGISNI 550
Query: 300 RIEDVTAINTESAIRIKSAVGRG 322
E V IN++ I +K+ GRG
Sbjct: 551 IAEQVHIINSKIGIELKTTKGRG 619
>AV769891
Length = 526
Score = 187 bits (474), Expect = 4e-48
Identities = 84/146 (57%), Positives = 109/146 (74%), Gaps = 5/146 (3%)
Frame = -3
Query: 316 KSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAK 375
K+AVGRG +VKDI+V+ M ++T+K+ FWMTG+YG H D +DPNALP+I GINYRD+ A
Sbjct: 524 KTAVGRGGYVKDIYVQRMTMHTIKWAFWMTGNYGSHADKNYDPNALPEIKGINYRDMVAD 345
Query: 376 NVTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELLQEK 435
VT+AG LEGISND FTGIC++NVTI M+A KK PW C+D+ G+TS V PKPC LL ++
Sbjct: 344 EVTMAGNLEGISNDQFTGICIANVTISMAAKSKKQPWTCSDVEGITSGVTPKPCNLLPDQ 165
Query: 436 EIE-----CPFPTDKLAIENVQFKTC 456
E C FPT+ L I+ ++ K C
Sbjct: 164 GPEKITTTCEFPTETLPIDMLELKKC 87
>TC18673 similar to UP|Q9LWY0 (Q9LWY0) ESTs AU029388(E30287), partial (23%)
Length = 560
Score = 184 bits (467), Expect = 3e-47
Identities = 90/179 (50%), Positives = 124/179 (68%)
Frame = +2
Query: 225 NTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSA 284
NTDGI+PDS +V IED +I +GDD IAIKSGWDEYGI G PS I+IRRL + S
Sbjct: 17 NTDGIDPDSSDDVCIEDCYISTGDDLIAIKSGWDEYGIAYGRPSTNIVIRRLVGKTQTSG 196
Query: 285 MVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYVFWM 344
+A+GSEMSGG+ +V ED+ ++ +AIRIK++ GRG +V++I++ M L ++Y
Sbjct: 197 -IAIGSEMSGGVSEVHAEDIHFYDSYNAIRIKTSRGRGGYVRNIYISNMTLVNIEYAITF 373
Query: 345 TGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEM 403
G YG+HPD+ +DPNALP I I +DV +N+ AG LEGI D F IC+SN+++ +
Sbjct: 374 NGLYGEHPDDAYDPNALPVIEKITIKDVIGENIKQAGILEGIEGDNFVNICLSNISLNV 550
>AV422277
Length = 454
Score = 177 bits (450), Expect = 3e-45
Identities = 78/149 (52%), Positives = 109/149 (72%)
Frame = +2
Query: 108 SQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFH 167
+Q+ WP + LPSYGRGR+ P GR+ S I G + DV+ITG+NGTIDGQG WW+ +
Sbjct: 8 TQELGNWPLIAPLPSYGRGRERPGGRYVSFIHGDGVRDVVITGENGTIDGQGDVWWNMWR 187
Query: 168 KKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTD 227
++ L TRP ++E + S I ISN+ +SP W +HP+Y SN+++ +TILAP DSPNTD
Sbjct: 188 QRTLQFTRPNLVEFVNSRNIIISNVIFKDSPFWNIHPVYCSNVVVRYVTILAPRDSPNTD 367
Query: 228 GINPDSCTNVRIEDNFIVSGDDCIAIKSG 256
G++PDS +NV IED++I +GDD +A+KSG
Sbjct: 368 GVDPDSSSNVCIEDSYISTGDDLVAVKSG 454
>TC19513 similar to UP|Q84LI7 (Q84LI7) Polygalacturonase-like protein,
partial (20%)
Length = 442
Score = 149 bits (375), Expect = 1e-36
Identities = 74/121 (61%), Positives = 92/121 (75%)
Frame = +2
Query: 1 VIGVISVVLILGSLAECRVPSSIKLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFN 60
V ++ V+L+ +AE R ++ +F Y AINCR HSA LTDFG +GDGKT NTKAF
Sbjct: 83 VYALLLVILLSSDVAESRKSKTVT-TSFVYSAINCRAHSASLTDFGGIGDGKTSNTKAFQ 259
Query: 61 AAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVL 120
+AI++LSQYA+ GGAQL VP GKWLTGSF+LTSHFTL+L KDAV+L SQ+ SEWP + L
Sbjct: 260 SAISHLSQYASKGGAQLYVPAGKWLTGSFSLTSHFTLYLNKDAVLLASQNISEWPVIEPL 439
Query: 121 P 121
P
Sbjct: 440 P 442
>AV423118
Length = 472
Score = 132 bits (333), Expect = 9e-32
Identities = 63/157 (40%), Positives = 97/157 (61%)
Frame = +2
Query: 195 INSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIK 254
+N+P + +HP+Y S + I+ LTI AP +SP+T G+ PDS +V IED I +G D IA+K
Sbjct: 2 LNAPAYSIHPVYCSYVHIHNLTIFAPPESPDTVGLVPDSSDHVCIEDCVIATGYDAIALK 181
Query: 255 SGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIR 314
SGWDEYGI G P++ + IRR+ + + +A GS+MSGGI +V +E+ N+ I
Sbjct: 182 SGWDEYGIAYGRPTENVHIRRVDLQASSGSALAFGSDMSGGISNVLVENAHLHNSNGGIE 361
Query: 315 IKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDH 351
++ GRG ++KDI + +++ + G G H
Sbjct: 362 FRTTRGRGGYMKDIIISDVEMKNVYTAIAARGHCGSH 472
>TC16334
Length = 710
Score = 101 bits (251), Expect = 3e-22
Identities = 44/110 (40%), Positives = 64/110 (58%)
Frame = +1
Query: 324 FVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKL 383
++KDI + L + M G YG HPD+ +DPNA+P + I +++V NV+IAG
Sbjct: 4 YMKDILISDTKLENIHLGIGMIGDYGSHPDDKYDPNAMPVVGSITFKNVVGGNVSIAGNF 183
Query: 384 EGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELLQ 433
GI + PF IC+SNVT +S+ W C+++ G + V PKPC LQ
Sbjct: 184 SGIVDSPFAPICLSNVTFSISSESSSPSWFCSNVMGFSEEVFPKPCSDLQ 333
>BP056102
Length = 472
Score = 87.0 bits (214), Expect = 6e-18
Identities = 37/93 (39%), Positives = 53/93 (56%)
Frame = -1
Query: 344 MTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEM 403
+ G GDHPD F+PNALP + GI +V V AG ++G+ N PFT +C+S + +
Sbjct: 433 IAGDVGDHPDEKFNPNALPVVKGITITNVWGVRVLQAGLIKGLKNSPFTDVCLSEINLHG 254
Query: 404 SAHKKKLPWNCTDISGVTSNVVPKPCELLQEKE 436
+ + PW C+D+SG V P PC L +E
Sbjct: 253 MSGPRSPPWKCSDVSGFAHQVSPWPCSQLSSQE 155
>TC20272 weakly similar to UP|Q9LRY8 (Q9LRY8) Polygalacturonase-like
protein, partial (5%)
Length = 494
Score = 64.7 bits (156), Expect = 3e-11
Identities = 28/86 (32%), Positives = 50/86 (57%)
Frame = +1
Query: 370 RDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPC 429
+D+T ++TIAG G+ + PFT IC+S++T+ M+ + W C+++SG + +V PKPC
Sbjct: 7 QDITGTDITIAGNFAGLQDSPFTNICLSHITLSMN-FASSITWECSNVSGYSDSVFPKPC 183
Query: 430 ELLQEKEIECPFPTDKLAIENVQFKT 455
L+ F L++ + +T
Sbjct: 184 PNLESHSDSSSFCLSMLSMPSTGRET 261
>AU088785
Length = 773
Score = 58.9 bits (141), Expect = 2e-09
Identities = 42/146 (28%), Positives = 69/146 (46%), Gaps = 4/146 (2%)
Frame = +2
Query: 176 PYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCT 235
P + M + + N+ +N + + +N+ + L ++AP SPNTDGI+ S
Sbjct: 14 PSSLFFMDXSNVVVQNIKSLNPKGFHMFVTKCTNVRLRKLKLIAPETSPNTDGIHISSSI 193
Query: 236 NVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGS----E 291
NV I N I +GDDCI++ G S+ + I RL C P +++GS
Sbjct: 194 NVIIARNTIQTGDDCISMIXG-----------SENVFINRLKC-GPGHG-ISIGSLGKYA 334
Query: 292 MSGGIQDVRIEDVTAINTESAIRIKS 317
++ + I + I T + +RIKS
Sbjct: 335 EXREVKGIXIXNSALIGTTNGLRIKS 412
>BP030312
Length = 418
Score = 50.8 bits (120), Expect = 5e-07
Identities = 24/40 (60%), Positives = 28/40 (70%)
Frame = +3
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPP 81
LTDFG VGDG TLNT AF A++ +S+ GG QL VPP
Sbjct: 297 LTDFGGVGDGVTLNTVAFERAVSAISKLREKGGGQLNVPP 416
>TC19633 similar to UP|Q7NET9 (Q7NET9) Glr3789 protein, partial (4%)
Length = 459
Score = 47.8 bits (112), Expect = 4e-06
Identities = 37/118 (31%), Positives = 57/118 (47%), Gaps = 1/118 (0%)
Frame = +3
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVP-PGKWLTGSFNLTSHFTLFLQ 100
++DFGA GDG +T + +AI + ++I P PGK+LT + L S LF++
Sbjct: 159 VSDFGAAGDGVRYDTTSIQSAINSCP---GSSPCRVIFPAPGKYLTATVFLKSGVVLFVE 329
Query: 101 KDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQ 158
A ILG ++P+ P+ + ++ N TDV I G G +DGQ
Sbjct: 330 PGATILGGPRLEDYPR------------EPSRWY--VVLAENATDVGIDG-GGVVDGQ 458
>AV770305
Length = 442
Score = 31.6 bits (70), Expect = 0.29
Identities = 17/46 (36%), Positives = 23/46 (49%)
Frame = +2
Query: 153 GTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSP 198
G IDG GS W++ T RP + + + +SNL L NSP
Sbjct: 317 GKIDGLGSSGWERCR----TCARPTSLSFYSGNGLTVSNLHLANSP 442
>TC12046
Length = 965
Score = 30.8 bits (68), Expect = 0.49
Identities = 18/43 (41%), Positives = 22/43 (50%), Gaps = 6/43 (13%)
Frame = -3
Query: 72 DGGAQLIVPPGK------WLTGSFNLTSHFTLFLQKDAVILGS 108
D G QL PGK WL F+L +F+QKD I+GS
Sbjct: 699 DYGVQLYKRPGKKGN*VPWLQQVFDLRQGSVVFIQKDYYIIGS 571
>TC10313 similar to UP|Q8LB12 (Q8LB12) Seh1-like protein, partial (34%)
Length = 774
Score = 28.1 bits (61), Expect = 3.2
Identities = 13/47 (27%), Positives = 24/47 (50%)
Frame = +2
Query: 228 GINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIR 274
G+NPD + +E ++SG D + + WD G+ + Q ++R
Sbjct: 143 GLNPDHDGRLPVERVAVLSGHDGMVWQMEWDMSGMTLATTGQDGMVR 283
>TC8653 similar to UP|AAS46232 (AAS46232) Methionine sulfoxide reductase A,
partial (71%)
Length = 1175
Score = 27.7 bits (60), Expect = 4.2
Identities = 14/33 (42%), Positives = 19/33 (57%)
Frame = +3
Query: 257 WDEYGIKVGMPSQQIIIRRLTCISPDSAMVALG 289
W G +G P Q+++I R +CI D M ALG
Sbjct: 414 WPSRGY-LG*PRQRLVIARASCIIQDMRMSALG 509
>TC12706 UP|Q7X9C0 (Q7X9C0) NIN-like protein 2, complete
Length = 3814
Score = 27.7 bits (60), Expect = 4.2
Identities = 21/77 (27%), Positives = 31/77 (39%), Gaps = 3/77 (3%)
Frame = +3
Query: 157 GQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPT---WFVHPIYSSNIIIN 213
G+ YW + +++ I SD + S L+N + WF P + I N
Sbjct: 378 GELGYWQSPSAQCGGSISSDNGISNSISDDMPNSFSELLNFDSYAGWFTGPSVTDQIFAN 557
Query: 214 GLTILAPVDSPNTDGIN 230
L A V P +DG N
Sbjct: 558 ELPSFASVSYPLSDGFN 608
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,686,013
Number of Sequences: 28460
Number of extensions: 98180
Number of successful extensions: 487
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 478
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 480
length of query: 463
length of database: 4,897,600
effective HSP length: 93
effective length of query: 370
effective length of database: 2,250,820
effective search space: 832803400
effective search space used: 832803400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC121237.2


