

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
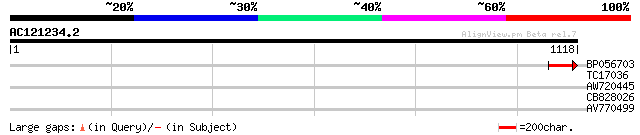
Query= AC121234.2 + phase: 0
(1118 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP056703 114 9e-26
TC17036 similar to UP|Q8VZD7 (Q8VZD7) AT3g28480/MFJ20_16, partia... 30 1.6
AW720445 28 8.0
CB828026 28 8.0
AV770499 28 8.0
>BP056703
Length = 386
Score = 114 bits (285), Expect = 9e-26
Identities = 50/57 (87%), Positives = 53/57 (92%)
Frame = -3
Query: 1062 VHLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFSVRLSPVTPATSGHDIYRSQLQNS 1118
+HLDHKHMGLGGDDSWSPCV DQYL+PPVPYSFSVRL PV P+TSGHDIYRSQL NS
Sbjct: 384 IHLDHKHMGLGGDDSWSPCVPDQYLIPPVPYSFSVRLCPVMPSTSGHDIYRSQLLNS 214
>TC17036 similar to UP|Q8VZD7 (Q8VZD7) AT3g28480/MFJ20_16, partial (21%)
Length = 697
Score = 30.4 bits (67), Expect = 1.6
Identities = 19/56 (33%), Positives = 26/56 (45%), Gaps = 4/56 (7%)
Frame = +2
Query: 1057 GDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPP----VPYSFSVRLSPVTPATSGH 1108
G+ I H + K + D+SWS C H Y V P FS+ L+ T + S H
Sbjct: 14 GETIFPHAESK-LSQPKDESWSECAHKGYAVKPRKGDALLFFSLHLNATTDSNSLH 178
>AW720445
Length = 500
Score = 28.1 bits (61), Expect = 8.0
Identities = 10/22 (45%), Positives = 17/22 (76%)
Frame = -3
Query: 939 KPNADLPPLPRVGIEMNLEKSL 960
+PN +P +P+V +EMNL+K +
Sbjct: 291 RPNKQVPAVPKV*LEMNLKKKI 226
>CB828026
Length = 542
Score = 28.1 bits (61), Expect = 8.0
Identities = 17/55 (30%), Positives = 22/55 (39%)
Frame = -1
Query: 85 WVKDLPFVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFD 139
W K P + F LAS PC P + + + S L PS+ L G D
Sbjct: 311 WTKTQPVFEVTVHS**FHLASQPCQ*PCCIQVQDLEGTSSSHLSFPSSLSLQGCD 147
>AV770499
Length = 491
Score = 28.1 bits (61), Expect = 8.0
Identities = 16/50 (32%), Positives = 27/50 (54%)
Frame = +2
Query: 67 SAVWKDDAVNGALESAAFWVKDLPFVKSLSGYWKFFLASNPCNVPAKFHD 116
S WK+ A +G+L+++ F F+ ++GY K + P N+ FHD
Sbjct: 302 SGNWKERAFSGSLQASPFLEMLYNFIHGVNGYGK----TEPINI-LGFHD 436
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,492,366
Number of Sequences: 28460
Number of extensions: 402387
Number of successful extensions: 1829
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 1813
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1827
length of query: 1118
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1018
effective length of database: 2,051,600
effective search space: 2088528800
effective search space used: 2088528800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC121234.2


