

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
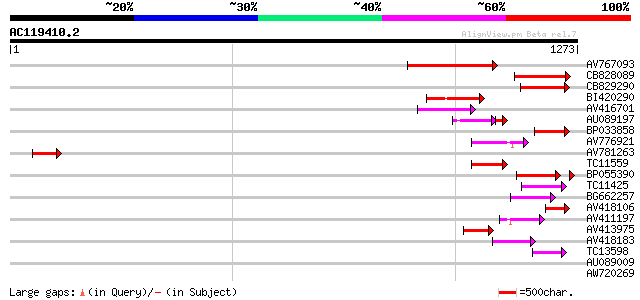
Query= AC119410.2 - phase: 1 /pseudo
(1273 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV767093 191 5e-49
CB828089 170 1e-42
CB829290 150 2e-36
BI420290 121 6e-28
AV416701 110 2e-24
AU089197 80 3e-21
BP033858 94 1e-19
AV776921 84 2e-16
AV781263 75 8e-14
TC11559 similar to UP|Q9SXT3 (Q9SXT3) Multidrug resistance prote... 67 1e-11
BP055390 67 3e-11
TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proeti... 62 4e-10
BG662257 60 2e-09
AV418106 57 2e-08
AV411197 56 3e-08
AV413975 52 5e-07
AV418183 49 7e-06
TC13598 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proeti... 47 2e-05
AU089009 40 0.002
AW720269 37 0.026
>AV767093
Length = 603
Score = 191 bits (486), Expect = 5e-49
Identities = 96/201 (47%), Positives = 130/201 (63%)
Frame = +1
Query: 894 RAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRIEALLNLTVITAALLLILLPQRY 953
R F F + + V+ + FH+N A EWL R++ + + + + +I LP
Sbjct: 1 RGFRKQGEFCQENIDRVNASLRMDFHNNGANEWLGYRLDFTGVVFLCISTMFMIFLPSSI 180
Query: 954 LSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVERIKQFIHIPAEPPAIVDNNRPP 1013
+ P VGLSLSY L L+ F N+ N ++SVERIKQF ++P+E P + + PP
Sbjct: 181 VRPEYVGLSLSYGLALSSVLSFTISMTCNVENKMVSVERIKQFTNLPSEAPWKIPDLSPP 360
Query: 1014 SSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLV 1073
+WP+ G I+L L+VRYRPN PLVLKGI+ T +GG ++GVVGRTGSGKSTLI LFRL+
Sbjct: 361 QNWPNHGSIELNSLQVRYRPNTPLVLKGISLTVQGGEKIGVVGRTGSGKSTLIQVLFRLI 540
Query: 1074 EPSRGDILIDGINICSMGLKD 1094
EPS G I+IDGINIC++GL D
Sbjct: 541 EPSAGKIIIDGINICTLGLHD 603
>CB828089
Length = 511
Score = 170 bits (430), Expect = 1e-42
Identities = 85/127 (66%), Positives = 100/127 (77%), Gaps = 1/127 (0%)
Frame = +1
Query: 1134 KCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSAT 1193
KCQL+E + + LDSSV ++G NWS+GQRQLFCLGR LL+R+RILVLDEATASID+AT
Sbjct: 7 KCQLQEAVQEKEGGLDSSVVEDGANWSMGQRQLFCLGRALLRRSRILVLDEATASIDNAT 186
Query: 1194 DAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSS-FSK 1252
D ILQ+ IR EF +CTVITVAHR+PTV+D V+ +S GKLVEYDEP LM S F K
Sbjct: 187 DLILQKTIRTEFADCTVITVAHRIPTVMDCTKVLAISDGKLVEYDEPMNLMKREGSLFGK 366
Query: 1253 LVAEYWS 1259
LV EYWS
Sbjct: 367 LVKEYWS 387
>CB829290
Length = 546
Score = 150 bits (378), Expect = 2e-36
Identities = 72/111 (64%), Positives = 96/111 (85%), Gaps = 1/111 (0%)
Frame = -3
Query: 1148 LDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEE 1207
LDS+VS+ G NWS+GQRQL CLGRVLLK++++LVLDEATAS+D+ATD ++Q+ +RQ F E
Sbjct: 532 LDSTVSENGENWSMGQRQLVCLGRVLLKKSKVLVLDEATASVDTATDNLIQQTLRQHFSE 353
Query: 1208 CTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLM-DTNSSFSKLVAEY 1257
TVIT+AHR+ +VIDSDMV++LS G + EYD P+KL+ D +SSF++LVAEY
Sbjct: 352 STVITIAHRITSVIDSDMVLLLSQGLIEEYDSPAKLLEDKSSSFAQLVAEY 200
>BI420290
Length = 398
Score = 121 bits (304), Expect = 6e-28
Identities = 59/133 (44%), Positives = 86/133 (64%), Gaps = 2/133 (1%)
Frame = +2
Query: 936 NLTVITAALLLILLPQRYLSPGRVGLSLSYALTLN--GAQIFWTRWFSNLSNYIISVERI 993
N + ++L+ LP+ ++P GL+++Y + LN A + W N N +ISVERI
Sbjct: 5 NFVFAFSLVMLVSLPEGIINPSIAGLAVTYGINLNVLQASVIWN--ICNAENKMISVERI 178
Query: 994 KQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRVG 1053
Q+ HI +E P ++++ +PPS+WP G I + L++RY + P VLK ITCTF G ++G
Sbjct: 179 LQYTHIASEAPLVIEDCKPPSNWPETGTICFKNLQIRYAEHLPSVLKNITCTFPGRKKIG 358
Query: 1054 VVGRTGSGKSTLI 1066
VVGRTGSGKSTLI
Sbjct: 359 VVGRTGSGKSTLI 397
>AV416701
Length = 406
Score = 110 bits (274), Expect = 2e-24
Identities = 55/130 (42%), Positives = 77/130 (58%)
Frame = +2
Query: 917 FFHSNVAMEWLVLRIEALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFW 976
FF S A+EWL LR+E L +LL+ P+ + P GL+ +Y L LN
Sbjct: 17 FFCSIAAIEWLCLRMELLSTFVFSFCMVLLVSFPRGNIDPSMAGLAATYGLNLNSRLSRR 196
Query: 977 TRWFSNLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAP 1036
F L N IIS+ERI Q+ +P+E P+++++ RP S+WP G I L L+VRY+ N P
Sbjct: 197 ILSFCKLENKIISIERIYQYSQVPSEAPSVIEDFRPTSTWPENGTIQLIDLKVRYKENLP 376
Query: 1037 LVLKGITCTF 1046
LVL G++CTF
Sbjct: 377 LVLHGVSCTF 406
>AU089197
Length = 658
Score = 79.7 bits (195), Expect(2) = 3e-21
Identities = 45/99 (45%), Positives = 59/99 (59%)
Frame = +1
Query: 995 QFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRVGV 1054
QF IP+E P + P G I+L L+V+Y P AP+VLK +T F G ++G+
Sbjct: 289 QFSSIPSEAPL----K*V*TXLPRDGMIELHKLQVQYDPAAPIVLKDVTFIFSGQKKIGI 456
Query: 1055 VGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLK 1093
RTGSGKSTL+ ALF +V P G ILIDG+ I + K
Sbjct: 457 XXRTGSGKSTLLQALF*IVXPLSGCILIDGVXIPRLACK 573
Score = 40.4 bits (93), Expect(2) = 3e-21
Identities = 17/28 (60%), Positives = 24/28 (85%)
Frame = +3
Query: 1090 MGLKDLRMKLSIIPQEPTLFKGSIRTNL 1117
+GL+DLR K+ IIPQ+PTLF ++RT+L
Sbjct: 561 IGLQDLRTKIGIIPQDPTLFLXTVRTDL 644
>BP033858
Length = 533
Score = 94.4 bits (233), Expect = 1e-19
Identities = 43/80 (53%), Positives = 60/80 (74%)
Frame = -1
Query: 1178 RILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEY 1237
R+L +DEATAS+DS TDA++QRIIR++F T+I++AHR+PTV+D D V+V+ G+ E+
Sbjct: 533 RLLFMDEATASVDSQTDAVIQRIIREDFAARTIISIAHRIPTVMDCDRVLVVDAGRAKEF 354
Query: 1238 DEPSKLMDTNSSFSKLVAEY 1257
D PS L+ S F LV EY
Sbjct: 353 DRPSNLLQRQSLFGALVQEY 294
>AV776921
Length = 435
Score = 83.6 bits (205), Expect = 2e-16
Identities = 51/133 (38%), Positives = 75/133 (56%), Gaps = 5/133 (3%)
Frame = +2
Query: 1038 VLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRM 1097
+L + G V +VG +GSGKST+IS + R EP GDIL+DG +I + LK LR
Sbjct: 41 ILNKLCLDIPSGKIVALVGGSGSGKSTVISLIERFYEPLSGDILLDGNDIRDLDLKWLRQ 220
Query: 1098 KLSIIPQEPTLFKGSIRTNLDPLGLYSDD-----EIWKAVEKCQLKETISKLPSLLDSSV 1152
++ ++ QEP LF SI+ N+ LY D E+ +AV+ + I+ LP L++ V
Sbjct: 221 QIGLVNQEPALFATSIKENI----LYGKDNATLEELKRAVKLSDAQSFINNLPERLETQV 388
Query: 1153 SDEGGNWSLGQRQ 1165
+ G S GQ+Q
Sbjct: 389 GERGIQLSGGQKQ 427
>AV781263
Length = 234
Score = 74.7 bits (182), Expect = 8e-14
Identities = 39/66 (59%), Positives = 49/66 (74%)
Frame = +1
Query: 51 TSS*RHSFPCF*R*SKHGL*KICSCLGITC*RED*EQYKEFGSLVYS*IIPKRKHINSIL 110
+SS*RH FPCF R S+ L K+C+ +GI C E+ EQY+E SLVYS + KRKHINSIL
Sbjct: 34 SSS*RHPFPCFRRRSRLCLPKLCAFMGIPCEGEEQEQYQELVSLVYSQNLLKRKHINSIL 213
Query: 111 CINQNY 116
C+ QN+
Sbjct: 214 CVAQNH 231
>TC11559 similar to UP|Q9SXT3 (Q9SXT3) Multidrug resistance protein
(Fragment), partial (41%)
Length = 460
Score = 67.4 bits (163), Expect = 1e-11
Identities = 34/80 (42%), Positives = 50/80 (62%)
Frame = +3
Query: 1038 VLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRM 1097
+LKGI G VGV+G +GSGKSTL+ AL RL EP + +D +IC + + LR
Sbjct: 135 ILKGIHLDIPKGVIVGVIGPSGSGKSTLLRALNRLWEPPSASVFLDARDICHLDVLSLRR 314
Query: 1098 KLSIIPQEPTLFKGSIRTNL 1117
K+ ++ Q P LF+G++ N+
Sbjct: 315 KVGMLFQLPALFEGTVADNV 374
>BP055390
Length = 488
Score = 66.6 bits (161), Expect(2) = 3e-11
Identities = 34/99 (34%), Positives = 63/99 (63%)
Frame = +3
Query: 1138 KETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAIL 1197
K++++ +LL + V G S GQ+Q + R +LK IL+LDEAT+++D+ ++ ++
Sbjct: 171 KDSVN*CSNLL-TQVGQRGVQLSGGQKQRIAIARAILKNPPILLLDEATSALDTESEKLV 347
Query: 1198 QRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1236
Q + TVI +AHR+ TV+++D++ V+ G++VE
Sbjct: 348 QEALETAMHGRTVILIAHRLSTVVNADVIAVVENGQVVE 464
Score = 19.6 bits (39), Expect(2) = 3e-11
Identities = 5/11 (45%), Positives = 8/11 (72%)
Frame = +2
Query: 1258 WSSCRKNSLPY 1268
W+SCR + P+
Sbjct: 449 WTSCRNRNTPH 481
>TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proetin, partial
(9%)
Length = 712
Score = 62.4 bits (150), Expect = 4e-10
Identities = 31/102 (30%), Positives = 59/102 (57%)
Frame = +2
Query: 1149 DSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEEC 1208
D+ V + G S GQ+Q + R ++K IL+LDEAT+++D+ ++ ++Q + +
Sbjct: 77 DTVVGERGTLLSGGQKQRVAIARAIIKTPNILLLDEATSALDAESERVVQDALDKVMVNR 256
Query: 1209 TVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSF 1250
T + VAHR+ T+ ++D++ VL G +VE L++ +
Sbjct: 257 TTVIVAHRLSTIKNADVITVLKNGVIVEKGRHETLINIKDGY 382
>BG662257
Length = 336
Score = 60.5 bits (145), Expect = 2e-09
Identities = 34/101 (33%), Positives = 58/101 (56%)
Frame = +1
Query: 1124 SDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLD 1183
++ EI A E + IS L D+ V D G S GQ+Q + R ++K +IL+LD
Sbjct: 34 TEAEIIAAAELANAHKFISSLQQGYDTVVGDRGIQLSGGQKQRVAIARAIVKSPKILLLD 213
Query: 1184 EATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSD 1224
EAT+++D+ ++ ++Q + + + T I VAHR+ T+ +D
Sbjct: 214 EATSALDAESEKVVQDALDRVRVDRTTIVVAHRLSTIKGAD 336
>AV418106
Length = 419
Score = 57.0 bits (136), Expect = 2e-08
Identities = 23/55 (41%), Positives = 40/55 (71%)
Frame = -2
Query: 1203 QEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEY 1257
++F + T++++AHR+PTV+D D V+V+ G E+D+PS+L++ + F LV EY
Sbjct: 418 EDFADRTIVSIAHRIPTVMDCDRVLVIDAGFAKEFDKPSRLLERPALFGALVKEY 254
>AV411197
Length = 391
Score = 56.2 bits (134), Expect = 3e-08
Identities = 36/106 (33%), Positives = 58/106 (53%), Gaps = 5/106 (4%)
Frame = +3
Query: 1101 IIPQEPTLFKGSIRTNLDPLGLY-----SDDEIWKAVEKCQLKETISKLPSLLDSSVSDE 1155
+I QEP +F +IR N+ +Y S+ E+ +A IS LP D+ V
Sbjct: 54 LIQQEPIIFSTTIRENI----IYARHNASEAEMKEAARIANAHHFISSLPHGYDTHVGMR 221
Query: 1156 GGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRII 1201
G + + GQ+Q + RV+LK IL+LDEA++SI+S + ++Q +
Sbjct: 222 GVDLTPGQKQRIAIARVVLKNAPILLLDEASSSIESESSRVIQEAL 359
>AV413975
Length = 343
Score = 52.4 bits (124), Expect = 5e-07
Identities = 27/68 (39%), Positives = 45/68 (65%), Gaps = 2/68 (2%)
Frame = +2
Query: 1020 GKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSR 1077
G+++L + Y RP+ P VLKGIT GS+V +VG +G GK+T+ + + R +P++
Sbjct: 143 GEVELDDVWFSYPSRPSHP-VLKGITMKLHPGSKVALVGPSGGGKTTIANLIERFYDPTK 319
Query: 1078 GDILIDGI 1085
G IL++G+
Sbjct: 320 GKILLNGV 343
>AV418183
Length = 298
Score = 48.5 bits (114), Expect = 7e-06
Identities = 32/98 (32%), Positives = 47/98 (47%), Gaps = 2/98 (2%)
Frame = +3
Query: 1084 GINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNL--DPLGLYSDDEIWKAVEKCQLKETI 1141
G+ + + + L K+SI+ QEPTLF SI N+ G + +I A + E I
Sbjct: 3 GVPLAEISHRHLHRKISIVSQEPTLFNCSIEENIAYGFDGKLNSVDIENAAKMANAHEFI 182
Query: 1142 SKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRI 1179
SK P + V + G S GQ+Q + R LL +I
Sbjct: 183 SKFPEKYQTFVGERGVRLSGGQKQRIAIARALLMDPKI 296
>TC13598 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proetin, partial
(7%)
Length = 552
Score = 46.6 bits (109), Expect = 2e-05
Identities = 22/77 (28%), Positives = 43/77 (55%)
Frame = +3
Query: 1174 LKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGK 1233
+K IL+LDE T+++D ++ ++Q + + T + VAHR+ T+ +D+++VL G
Sbjct: 3 IKSPNILLLDEXTSALDVESERVVQDALDKVMVNRTTVIVAHRLSTIKSADVIIVLKNGV 182
Query: 1234 LVEYDEPSKLMDTNSSF 1250
+VE L+ +
Sbjct: 183 IVEKGRHETLISIKDGY 233
>AU089009
Length = 209
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/54 (46%), Positives = 31/54 (57%)
Frame = +3
Query: 532 PR*SYRNR*ERD*HEWRTKAKDSTS*SSLQ*C*YLSS**SFQCS*CSYSCNTIQ 585
P * + NR*+R +WRTKA ST SL C* +S *S C *C+Y +Q
Sbjct: 9 PW*PH*NR*KRSKPQWRTKAASSTRPCSLSKC*CISFG*SXXCC*CAYCFKFVQ 170
>AW720269
Length = 524
Score = 36.6 bits (83), Expect = 0.026
Identities = 48/174 (27%), Positives = 74/174 (41%), Gaps = 16/174 (9%)
Frame = +3
Query: 1032 RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSM- 1090
+P +LK ++ + V VVG +G+GKSTL+ + V+ D IN M
Sbjct: 36 KPKPVNILKSVSFVARSSEIVAVVGPSGTGKSTLLRIIAGRVKDRDFDPKTISINDHPMT 215
Query: 1091 GLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETI-----SKLP 1145
LR + QE NL PL + ++ A K +LKE ++
Sbjct: 216 SPAQLRKICGFVAQE---------DNLLPLLTVKETLLFSA--KFRLKEMTPNDREMRVE 362
Query: 1146 SLL---------DSSVSDEGGNW-SLGQRQLFCLGRVLLKRNRILVLDEATASI 1189
SL+ DS V DE S G+R+ +G ++ IL+LDE T+ +
Sbjct: 363 SLMQELGLFHVSDSFVGDEENRGISGGERKRVSIGVDMIHNPPILLLDEPTSGL 524
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.346 0.150 0.508
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,672,175
Number of Sequences: 28460
Number of extensions: 366314
Number of successful extensions: 5092
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 4860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5070
length of query: 1273
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1172
effective length of database: 2,023,140
effective search space: 2371120080
effective search space used: 2371120080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC119410.2


