

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.8 + phase: 0
(908 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
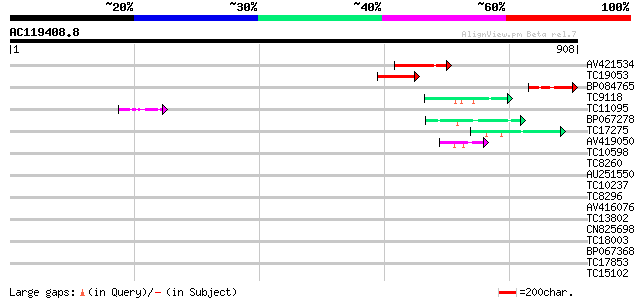
Score E
Sequences producing significant alignments: (bits) Value
AV421534 141 5e-34
TC19053 similar to UP|Q9FHH9 (Q9FHH9) DNA-binding protein-like, ... 113 1e-25
BP084765 94 1e-19
TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding l... 47 1e-05
TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 42 6e-04
BP067278 42 6e-04
TC17275 42 6e-04
AV419050 41 7e-04
TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526 At2g01... 40 0.001
TC8260 similar to UP|Q94ES8 (Q94ES8) Nodule extensin (Fragment),... 39 0.004
AU251550 39 0.004
TC10237 similar to UP|NO75_SOYBN (P08297) Early nodulin 75 precu... 39 0.004
TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1... 39 0.004
AV416076 38 0.006
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 37 0.018
CN825698 37 0.018
TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 37 0.018
BP067368 36 0.024
TC17853 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, ... 36 0.031
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 35 0.041
>AV421534
Length = 270
Score = 141 bits (355), Expect = 5e-34
Identities = 69/92 (75%), Positives = 79/92 (85%), Gaps = 1/92 (1%)
Frame = +3
Query: 617 DARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERP 676
D RRKRENERR ERDFSK+R+FGRE SSVGDVSS+K KTKP PP +SDYH +RHRSERP
Sbjct: 3 DLRRKRENERRGERDFSKEREFGREASSVGDVSSMKPKTKPFPPPPSSDYHHHRHRSERP 182
Query: 677 SPDSSHREVEPPRPTKRKSDH-SEREREDRDR 707
SPD REVEPPRP KRKS+H S+R+R+D+DR
Sbjct: 183 SPD---REVEPPRPAKRKSEHISDRQRDDKDR 269
>TC19053 similar to UP|Q9FHH9 (Q9FHH9) DNA-binding protein-like, partial
(3%)
Length = 603
Score = 113 bits (283), Expect = 1e-25
Identities = 57/69 (82%), Positives = 62/69 (89%), Gaps = 2/69 (2%)
Frame = +1
Query: 589 HRDLAEFSMGMNVPPP--AMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVG 646
HRDLA+FSMGMNVPPP MSREEFEARKAD RRKRENERR ERDFS++R+FGRE SSVG
Sbjct: 1 HRDLADFSMGMNVPPPPPVMSREEFEARKADLRRKRENERRGERDFSQEREFGREASSVG 180
Query: 647 DVSSIKSKT 655
DVSS+K KT
Sbjct: 181 DVSSMKPKT 207
>BP084765
Length = 451
Score = 94.0 bits (232), Expect = 1e-19
Identities = 49/78 (62%), Positives = 56/78 (70%), Gaps = 1/78 (1%)
Frame = -1
Query: 832 AATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPPPPVDD 891
+A S +A A + + GRKS V+ DYESSDD ERHFKRRPSRYEPSPPPP D
Sbjct: 451 SAVGSTAASASANGYYQ--GRKSSKVVPDYESSDD----ERHFKRRPSRYEPSPPPPAAD 290
Query: 892 WVEE-GRHSRGTRDRKHR 908
W EE GRHSRG+R+RKHR
Sbjct: 289 WEEEGGRHSRGSRERKHR 236
>TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding
like-protein, partial (16%)
Length = 779
Score = 47.0 bits (110), Expect = 1e-05
Identities = 42/169 (24%), Positives = 63/169 (36%), Gaps = 27/169 (15%)
Frame = +2
Query: 664 SDYHQNRHRSERPSPDSSHREVEPPRPTKRK--SDHSEREREDRDRDYDYH--------D 713
S H+ E + +V + +++K S S DRDRD H D
Sbjct: 5 SSRHEEEEEEEEEEEEEKREDVRDHKRSRKKHRSHRSSHTSRDRDRDGRKHKRRHSSSKD 184
Query: 714 RHQDRDYXH----DRHQHRRHHHRTEASSKKS-------------TDPVTKSTSRKSSEP 756
++ R H D HQH HHH+ ++SS + +D K + R E
Sbjct: 185 KYSYRGAKHGISDDEHQHSSHHHKYDSSSDEKHRSSRRRQREDSMSDHEHKHSRRHKHET 364
Query: 757 DIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRIS 805
+ + R S TK S S + E +D+ Q+AS R S
Sbjct: 365 SSEDERRHRSRH-TKHKSRSHVERESELEEGEVMKSDKSQQASEVERAS 508
>TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (89%)
Length = 912
Score = 41.6 bits (96), Expect = 6e-04
Identities = 28/78 (35%), Positives = 35/78 (43%)
Frame = +1
Query: 175 LDWQHQGSDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNG 234
++ H G G G GRG GGR GGG G G + K C+ C PGHF + C G
Sbjct: 280 VELSHNSRGGGGGGGGGRG-GGRSGGGGG-GSDMK-------CYECGEPGHFARECRMRG 432
Query: 235 DPNYDVKRVKQPTGIPRS 252
+R + P RS
Sbjct: 433 GSGR--RRSRSPPRFRRS 480
>BP067278
Length = 562
Score = 41.6 bits (96), Expect = 6e-04
Identities = 37/165 (22%), Positives = 61/165 (36%), Gaps = 6/165 (3%)
Frame = +1
Query: 667 HQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRH------QDRDY 720
H+ R RS S D + R + RK D S + R R D+H ++
Sbjct: 46 HRERKRSRSVSVDEKPQRRS--RSSPRKVDESGSRHKKRSRSKSVEDKHNFPEKLEENRI 219
Query: 721 XHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAP 780
RH +RH S+ TD +R+ KSK R S +KS+ +
Sbjct: 220 RKSRHNDKRHSRSRSTESRDQTD------AREDERKYGKSKHRDSKRTRSKSVEGKHRSK 381
Query: 781 TTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASS 825
S E + R ++ S +S + + + ++ L S+
Sbjct: 382 DKSGEVKDKKSKHRDRRRS--RSVSLEGDHDIGGTSSRKNLDESN 510
>TC17275
Length = 796
Score = 41.6 bits (96), Expect = 6e-04
Identities = 40/161 (24%), Positives = 64/161 (38%), Gaps = 9/161 (5%)
Frame = +2
Query: 739 KKSTDPVTKSTSRKSSEPDIKSK----SRKSSEPVTKSLSLSKTAPTTSAEA-----AAA 789
KK T P+T + S K+ IKS S K+ T + S K +TS + + +
Sbjct: 209 KKPTKPITTNLSSKNQTKSIKSNNNNLSSKNQTKTTLNASTFKKLNSTSIKIKKLNNSTS 388
Query: 790 AAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSN 849
+ K S S+ + A A AK + + +TT T V+ L+
Sbjct: 389 KSFTNSSKTSTTSKKQLDLTKLAGAIAKNK--TTKATTATDTKNKIKVNPGHIEEEKLNK 562
Query: 850 GGRKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPPPPVD 890
K K ++ D+D D FK P R++ S P ++
Sbjct: 563 QSNKKKQSPPNWMFEDEDSDLVSDFKDLPIRFQKSLIPDLE 685
>AV419050
Length = 409
Score = 41.2 bits (95), Expect = 7e-04
Identities = 31/94 (32%), Positives = 39/94 (40%), Gaps = 15/94 (15%)
Frame = +1
Query: 689 RPTKRKSDHSEREREDRDRDYDY------HDRHQDRDYXHDRH---------QHRRHHHR 733
R +R+ + ER+R DRDRD D R + D+ RH HRR HHR
Sbjct: 25 RDRERRREKEERDRGDRDRDRDRTRSKRSRSRSRSIDHGRSRHARSRSPSERSHRRRHHR 204
Query: 734 TEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSE 767
T S D K R S E D K + S+
Sbjct: 205 T-----PSPDQPRKRHRRDSVEDDHKDTKKVVSD 291
Score = 32.3 bits (72), Expect = 0.34
Identities = 28/92 (30%), Positives = 44/92 (47%), Gaps = 1/92 (1%)
Frame = +1
Query: 614 RKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRS 673
R + RR++E R +RD +DRD R S +S+++ I S + ++R S
Sbjct: 25 RDRERRREKEERDRGDRD--RDRDRTRSKRS-------RSRSRSI-DHGRSRHARSRSPS 174
Query: 674 ERPSPDSSHREVEPPRPTKR-KSDHSERERED 704
ER HR P +P KR + D E + +D
Sbjct: 175 ERSHRRRHHRTPSPDQPRKRHRRDSVEDDHKD 270
>TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526
At2g01100/F23H14.7 {Arabidopsis thaliana;}, partial
(33%)
Length = 902
Score = 40.4 bits (93), Expect = 0.001
Identities = 33/126 (26%), Positives = 49/126 (38%), Gaps = 1/126 (0%)
Frame = +3
Query: 607 SREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDY 666
S +E E+R + RR++ +++ RD D SS +VS K + K
Sbjct: 573 SGDESESRSEEERRRKRKQKKKLRDQVSRSDSSDSDSSADEVSKRKRQHK---------- 722
Query: 667 HQNRHRSERPS-PDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRH 725
RHR + S D S E P + H +R R + D D H +
Sbjct: 723 ---RHRVTKSSGSDFSSDEENSPVRKRSHGKHHKRHRHSESSESDLSS-----DDIHHKK 878
Query: 726 QHRRHH 731
HR+HH
Sbjct: 879 SHRKHH 896
Score = 37.7 bits (86), Expect = 0.008
Identities = 30/139 (21%), Positives = 58/139 (41%), Gaps = 3/139 (2%)
Frame = +3
Query: 617 DARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERP 676
+ R+++ ++R+ ++ S+ DF + S + K K + + R +
Sbjct: 507 EKRKEKLSKRKTKKWSSESSDFSGDESESRSEEERRRKRK----------QKKKLRDQVS 656
Query: 677 SPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQD---RDYXHDRHQHRRHHHR 733
DSS + +KRK H +R R + D+ ++ R H +H H+RH H
Sbjct: 657 RSDSSDSDSSADEVSKRKRQH-KRHRVTKSSGSDFSSDEENSPVRKRSHGKH-HKRHRHS 830
Query: 734 TEASSKKSTDPVTKSTSRK 752
+ S S+D + S +
Sbjct: 831 ESSESDLSSDDIHHKKSHR 887
Score = 32.3 bits (72), Expect = 0.34
Identities = 40/163 (24%), Positives = 63/163 (38%), Gaps = 4/163 (2%)
Frame = +3
Query: 607 SREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDY 666
S ++++ R R + R+ ++ D D + + K KTK S +SD+
Sbjct: 411 SESDYDSDDESKRTSRRSHRKHKKRSHYDSDHEKRKEKLS-----KRKTKKWS-SESSDF 572
Query: 667 HQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQ 726
S D S E R KRK R++ R D D D + + Q
Sbjct: 573 ----------SGDESESRSEEERRRKRKQKKKLRDQVSRSDSSD-SDSSAD-EVSKRKRQ 716
Query: 727 HRRHHHRTEASSKKSTD----PVTKSTSRKSSEPDIKSKSRKS 765
H+RH + S S+D PV K + K + S+S +S
Sbjct: 717 HKRHRVTKSSGSDFSSDEENSPVRKRSHGKHHKRHRHSESSES 845
>TC8260 similar to UP|Q94ES8 (Q94ES8) Nodule extensin (Fragment), partial
(79%)
Length = 1017
Score = 38.9 bits (89), Expect = 0.004
Identities = 26/89 (29%), Positives = 36/89 (40%)
Frame = +3
Query: 502 PHDLGAENYMMQMGPPPGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMP 561
P ++ A +Y PPP ++ P + PY P + PY+ P P +
Sbjct: 84 PSEISANHYAYSSPPPPPKPYSYHSPPPPVHSPPPPYEKPHPVYHSPPPPYEKPHP--VY 257
Query: 562 HDPFANGMPHDPFGMQGYMMPPIPPPPHR 590
H P PH P Y P PPPPH+
Sbjct: 258 HSP-PPPPPHKP-----YKYPSPPPPPHK 326
>AU251550
Length = 329
Score = 38.9 bits (89), Expect = 0.004
Identities = 32/92 (34%), Positives = 35/92 (37%), Gaps = 1/92 (1%)
Frame = +2
Query: 516 PPPGY-NPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPF 574
PPPGY NP P GP Y + P P G P DP+A P PF
Sbjct: 71 PPPGYHNP--------------PPPGPPQPPGYYNPP-----PPGPPRDPYAPAPPPPPF 193
Query: 575 GMQGYMMPPIPPPPHRDLAEFSMGMNVPPPAM 606
G PPPPH + PPPAM
Sbjct: 194 GHHH------PPPPHGP-------PSPPPPAM 250
>TC10237 similar to UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75)
(NGM-75), partial (49%)
Length = 803
Score = 38.9 bits (89), Expect = 0.004
Identities = 34/110 (30%), Positives = 43/110 (38%), Gaps = 5/110 (4%)
Frame = +1
Query: 499 WKPPHDLGAENYMM-QMGPPPGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFP 557
+ PP++ Y Q PPP Y+P + G PY H+ H PY+ P P
Sbjct: 211 YPPPYEKPPPEYEPPQEKPPPVYSPPYERPPT---GHPTPYPPFYHLPPVYHPPYEKPPP 381
Query: 558 NGMPHDPFANGMPHD--PFGMQGYMMPPIPPPPHR--DLAEFSMGMNVPP 603
P PH+ P + PPI PPH L E G N PP
Sbjct: 382 LYQP--------PHEKPPIYEPPHEKPPIYEPPHEKPPLYEPPPGYNPPP 507
>TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;}, partial (55%)
Length = 1276
Score = 38.9 bits (89), Expect = 0.004
Identities = 32/121 (26%), Positives = 49/121 (40%), Gaps = 4/121 (3%)
Frame = +2
Query: 679 DSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXH--DRHQHR-RHHHRTE 735
+S R + R + H + R RDRDY R + D DRH+ R R + R
Sbjct: 407 ESEERAKQGSRSRSPRRRHRDDHRSSRDRDYRRRSRSRSYDKYERGDRHRGRDRDYRRRS 586
Query: 736 ASSKKSTDPVTKSTSRKSSEPD-IKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADR 794
S S D + R EP +S+S S P +S S ++ + + + + R
Sbjct: 587 RSRSASLDYKGRGRGRYDDEPSRSRSRSVDSRSPARRSPSPRRSPSPKRSVSPKRSTSPR 766
Query: 795 K 795
K
Sbjct: 767 K 769
>AV416076
Length = 430
Score = 38.1 bits (87), Expect = 0.006
Identities = 25/102 (24%), Positives = 35/102 (33%)
Frame = +2
Query: 654 KTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHD 713
K KP PP + S H N H SP + H PP P R
Sbjct: 101 KAKPPPPRAFSSAHANHHHLHASSPRTHHHHR*PPPPLLRSPASMAATTA---------- 250
Query: 714 RHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSE 755
H +H RRH + + ++ P S R+++E
Sbjct: 251 ------ITHSQHHERRHQRKDHSENETLKPPPFFSDLRRTTE 358
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 36.6 bits (83), Expect = 0.018
Identities = 33/114 (28%), Positives = 54/114 (46%), Gaps = 1/114 (0%)
Frame = +2
Query: 733 RTEASSKKSTDPVTKSTSRKSSEPDIKSKSRK-SSEPVTKSLSLSKTAPTTSAEAAAAAA 791
R +SS S P + T KS +P S S SS P+T S + S+++PT ++ +++A+
Sbjct: 191 RAPSSSPASASPASSPT--KSPKPSSPSASAPPSSTPLTPSTATSESSPTATSSSSSASP 364
Query: 792 ADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTT 845
A P+ A + A + K ASS + S+ + SA ST+
Sbjct: 365 AP-------------PTSFSALSRALELKALASSPSPPSSPPPSPPSAICMSTS 487
Score = 29.6 bits (65), Expect = 2.2
Identities = 17/47 (36%), Positives = 20/47 (42%), Gaps = 6/47 (12%)
Frame = +3
Query: 657 PIPPSSASDYHQNRHRSERPSPDSS---HREVEPPRPTK---RKSDH 697
P PP H+N H RP P HR PP P + R+S H
Sbjct: 285 PQPP*RPPRRHRNPHPPRRPRPPQQVRRHRRASPPCPVR*S*RRSPH 425
>CN825698
Length = 704
Score = 36.6 bits (83), Expect = 0.018
Identities = 36/122 (29%), Positives = 43/122 (34%), Gaps = 3/122 (2%)
Frame = +1
Query: 582 PPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKD---RDF 638
PP PPPP L S PPPA R +RR +RDF + RD+
Sbjct: 139 PPPPPPPPPSLPPPSDSDLPPPPA----------------RRRDRRDDRDFDRPPNRRDY 270
Query: 639 GREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHS 698
+S P P D+ + R S PS R PP P KR S
Sbjct: 271 -----------YDRSNNSPPPKDRDRDFKRRRSPSP-PSQRDRDRRHSPPPPYKRSRRGS 414
Query: 699 ER 700
R
Sbjct: 415 PR 420
>TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (42%)
Length = 587
Score = 36.6 bits (83), Expect = 0.018
Identities = 18/43 (41%), Positives = 22/43 (50%)
Frame = +1
Query: 192 RGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNG 234
+G GG GGGRG G E C+ C PGHF + C + G
Sbjct: 109 KGGGGGRGGGRGRGGE------DLKCYECGEPGHFARECRSRG 219
>BP067368
Length = 322
Score = 36.2 bits (82), Expect = 0.024
Identities = 30/112 (26%), Positives = 44/112 (38%), Gaps = 4/112 (3%)
Frame = -2
Query: 612 EARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRH 671
E K+ R E R +R+ +DR+ RE +K K + ++R
Sbjct: 297 EQHKSAPSRSEEPRVREDRNVDRDREKSRE--------RVKDKD--------GERERSRE 166
Query: 672 RSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDY----DYHDRHQDRD 719
RS + D HRE R R D +RDRD D DR ++R+
Sbjct: 165 RSSDRARDRDHREDRHHRDRDRTRDRDRDRERERDRDVTEIEDVTDRGRERE 10
Score = 30.8 bits (68), Expect = 1.00
Identities = 29/82 (35%), Positives = 35/82 (42%), Gaps = 10/82 (12%)
Frame = -2
Query: 672 RSERPSPDSSH---REVEPPRP-TKRKSDHSERERE---DRDRDYDYHD--RHQDRDYXH 722
RSE P R+ E R K K ER RE DR RD D+ + H+DRD
Sbjct: 273 RSEEPRVREDRNVDRDREKSRERVKDKDGERERSRERSSDRARDRDHREDRHHRDRDRTR 94
Query: 723 DRHQHR-RHHHRTEASSKKSTD 743
DR + R R R + TD
Sbjct: 93 DRDRDRERERDRDVTEIEDVTD 28
Score = 28.9 bits (63), Expect = 3.8
Identities = 19/49 (38%), Positives = 25/49 (50%)
Frame = -2
Query: 607 SREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKT 655
SRE R D R RE+ +RD ++DRD RE DV+ I+ T
Sbjct: 174 SRERSSDRARD-RDHREDRHHRDRDRTRDRDRDRERERDRDVTEIEDVT 31
>TC17853 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, partial
(58%)
Length = 881
Score = 35.8 bits (81), Expect = 0.031
Identities = 18/48 (37%), Positives = 24/48 (49%)
Frame = +2
Query: 188 RGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGD 235
RG R G RG+G R + G C +C PGHF + CP+ G+
Sbjct: 338 RGRDRDDRGDRDRSRGYGGSRGS--NGGECFKCGKPGHFARECPSEGE 475
Score = 27.7 bits (60), Expect = 8.5
Identities = 10/21 (47%), Positives = 10/21 (47%)
Frame = -1
Query: 712 HDRHQDRDYXHDRHQHRRHHH 732
HD Y HDRH H H H
Sbjct: 401 HDYLHSLGYDHDRHDHHDHDH 339
Score = 27.7 bits (60), Expect = 8.5
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = +2
Query: 692 KRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHR 728
K + S R+R+D DR Y +R +DRD DR + R
Sbjct: 281 KAQPQGSSRDRDDGDR---YRERGRDRDDRGDRDRSR 382
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 35.4 bits (80), Expect = 0.041
Identities = 48/199 (24%), Positives = 73/199 (36%), Gaps = 2/199 (1%)
Frame = +1
Query: 649 SSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRD 708
S+ K+ T PP S RS P + P P KS HS R
Sbjct: 193 STTKAATNSSPPES---------RSTTPPSSEPTCLLAPASPPTPKSSHSSTPPPSRSSS 345
Query: 709 YDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEP 768
++ A SK +P T S+ K S KSS P
Sbjct: 346 TTPKSKNATSLTAPSCPPPASPAATAPAPSKTPPNPPTNSS---------KPSSCKSSLP 498
Query: 769 VT--KSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASST 826
T SLS + +PTTS + ++ ++++ AS +A R S+SS+
Sbjct: 499 NTTPSSLSSAPPSPTTSPISTPSSPENQEKPAS------------TPPSAPPRSTSSSSS 642
Query: 827 TEASTAATASVSAKAPSTT 845
+ +T S + KAP+++
Sbjct: 643 SPINTPRKPSSATKAPASS 699
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.130 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,715,140
Number of Sequences: 28460
Number of extensions: 296400
Number of successful extensions: 3591
Number of sequences better than 10.0: 253
Number of HSP's better than 10.0 without gapping: 2891
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3386
length of query: 908
length of database: 4,897,600
effective HSP length: 99
effective length of query: 809
effective length of database: 2,080,060
effective search space: 1682768540
effective search space used: 1682768540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC119408.8


