

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.7 + phase: 0 /pseudo
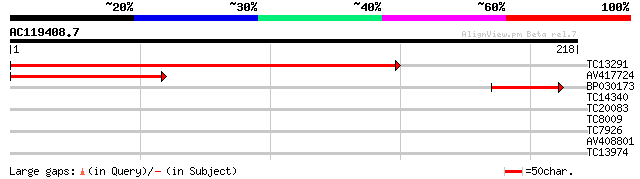
(218 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13291 277 9e-76
AV417724 114 1e-26
BP030173 57 2e-09
TC14340 homologue to UP|Q9ZW85 (Q9ZW85) 3-isopropylmalate dehydr... 28 0.93
TC20083 homologue to UP|Q8RVQ6 (Q8RVQ6) SEC6, partial (11%) 26 4.6
TC8009 homologue to UP|Q9LKJ2 (Q9LKJ2) Cytosolic phosphoglycerat... 26 4.6
TC7926 homologue to UP|PGKH_TOBAC (Q42961) Phosphoglycerate kina... 26 6.1
AV408801 25 7.9
TC13974 similar to UP|Q9SXE3 (Q9SXE3) T3P18.10, partial (14%) 25 7.9
>TC13291
Length = 564
Score = 277 bits (709), Expect = 9e-76
Identities = 136/150 (90%), Positives = 141/150 (93%)
Frame = +1
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQS QD IIPFQILGGESQVVQIMLKP EKIIAKPGSMCFMSGS+E
Sbjct: 115 MAAPFFSTPFQPYVYQSPQDEIIPFQILGGESQVVQIMLKPQEKIIAKPGSMCFMSGSIE 294
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
MENAYLPENEVG QWLFGKT+T+IV+ NSGPSDGFVGIAAPYFARILPIDLA FNG IL
Sbjct: 295 MENAYLPENEVGFRQWLFGKTITSIVLCNSGPSDGFVGIAAPYFARILPIDLAMFNGGIL 474
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVVAGA 150
CQPDAFLCSVNDVKVSNT DQRG+NVVAGA
Sbjct: 475 CQPDAFLCSVNDVKVSNTFDQRGQNVVAGA 564
>AV417724
Length = 315
Score = 114 bits (285), Expect = 1e-26
Identities = 55/60 (91%), Positives = 57/60 (94%)
Frame = +2
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQS QDAII FQILGGE+QVVQIMLKP EKIIAKPGSMCFMSGS+E
Sbjct: 134 MAAPFFSTPFQPYVYQSPQDAIIAFQILGGEAQVVQIMLKPEEKIIAKPGSMCFMSGSIE 313
>BP030173
Length = 498
Score = 57.4 bits (137), Expect = 2e-09
Identities = 26/28 (92%), Positives = 28/28 (99%)
Frame = -2
Query: 186 DVSCIVAVTSTVDIQIKYNGPARRTMFG 213
DVSCIVAVTSTV++QIKYNGPARRTMFG
Sbjct: 497 DVSCIVAVTSTVNVQIKYNGPARRTMFG 414
>TC14340 homologue to UP|Q9ZW85 (Q9ZW85) 3-isopropylmalate dehydratase,
small subunit, partial (16%)
Length = 548
Score = 28.5 bits (62), Expect = 0.93
Identities = 14/26 (53%), Positives = 18/26 (68%)
Frame = +1
Query: 55 MSGSVEMENAYLPENEVGIWQWLFGK 80
+S S+ MEN YLPE+ V + Q FGK
Sbjct: 142 VSVSIGMENFYLPEDHVDL*Q*RFGK 219
>TC20083 homologue to UP|Q8RVQ6 (Q8RVQ6) SEC6, partial (11%)
Length = 562
Score = 26.2 bits (56), Expect = 4.6
Identities = 14/43 (32%), Positives = 21/43 (48%)
Frame = -2
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGL 163
C+ + S + K+ + R NVV EVF +SG+GL
Sbjct: 174 CELATQIKSNQNAKIEESEISREENVVVRVEVFSATSVSGRGL 46
>TC8009 homologue to UP|Q9LKJ2 (Q9LKJ2) Cytosolic phosphoglycerate kinase ,
complete
Length = 1697
Score = 26.2 bits (56), Expect = 4.6
Identities = 11/24 (45%), Positives = 17/24 (70%)
Frame = +2
Query: 157 KLSGQGLAFILGGGSVVQKILEVG 180
+LSG+G+ I+GGG V + +VG
Sbjct: 1211 ELSGKGVTTIIGGGDSVAAVEKVG 1282
>TC7926 homologue to UP|PGKH_TOBAC (Q42961) Phosphoglycerate kinase,
chloroplast precursor , partial (86%)
Length = 1857
Score = 25.8 bits (55), Expect = 6.1
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +1
Query: 158 LSGQGLAFILGGGSVVQKILEVG 180
LSG+G+ I+GGG V + +VG
Sbjct: 1378 LSGKGVTTIIGGGDSVAAVEKVG 1446
>AV408801
Length = 308
Score = 25.4 bits (54), Expect = 7.9
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +2
Query: 157 KLSGQGLAFILGGGSVVQK 175
KL GQGL +ILG S QK
Sbjct: 107 KLEGQGLCYILGSVSCNQK 163
>TC13974 similar to UP|Q9SXE3 (Q9SXE3) T3P18.10, partial (14%)
Length = 528
Score = 25.4 bits (54), Expect = 7.9
Identities = 19/54 (35%), Positives = 26/54 (47%)
Frame = +2
Query: 21 AIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIW 74
A+ PFQ+ + V QI L P E + + S CF VEM+ Y E + W
Sbjct: 248 ALRPFQVDYKDWLVAQIGLPPLEDWMEQMYSECF-KNLVEMKENYRDEWDDNQW 406
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,428,752
Number of Sequences: 28460
Number of extensions: 40497
Number of successful extensions: 178
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 178
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 178
length of query: 218
length of database: 4,897,600
effective HSP length: 87
effective length of query: 131
effective length of database: 2,421,580
effective search space: 317226980
effective search space used: 317226980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC119408.7


