

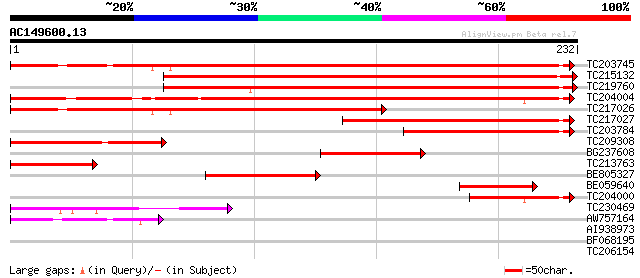
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149600.13 + phase: 0 /pseudo
(232 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203745 weakly similar to UP|O88972 (O88972) KH domain RNA bind... 309 8e-85
TC215132 similar to UP|Q75GR5 (Q75GR5) Expressed protein, partia... 284 3e-77
TC219760 weakly similar to UP|O88972 (O88972) KH domain RNA bind... 264 3e-71
TC204004 similar to UP|Q8W574 (Q8W574) At1g09660/F21M12_5, parti... 204 3e-53
TC217026 weakly similar to UP|O88972 (O88972) KH domain RNA bind... 203 6e-53
TC217027 weakly similar to UP|O88972 (O88972) KH domain RNA bind... 154 3e-38
TC203784 105 2e-23
TC209308 similar to UP|Q75GR5 (Q75GR5) Expressed protein, partia... 87 5e-18
BG237608 homologue to PIR|T01782|T017 GDP dissociation inhibitor... 73 1e-13
TC213763 similar to UP|Q75GR5 (Q75GR5) Expressed protein, partia... 70 8e-13
BE805327 similar to GP|20161243|dbj P0408G07.16 {Oryza sativa (j... 60 8e-10
BE059640 54 8e-08
TC204000 homologue to UP|Q84XC3 (Q84XC3) Ubiquitin/ribosomal fus... 48 4e-06
TC230469 similar to GB|BAB09296.1|9758634|AB011476 RNA-binding p... 41 4e-04
AW757164 41 5e-04
AI938973 31 0.55
BF068195 29 2.1
TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division prote... 27 7.9
>TC203745 weakly similar to UP|O88972 (O88972) KH domain RNA binding protein
QKI-7B, partial (20%)
Length = 1338
Score = 309 bits (791), Expect = 8e-85
Identities = 168/242 (69%), Positives = 193/242 (79%), Gaps = 11/242 (4%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTG--- 57
MQ LP+C+RLLNQEILR SG ++ NQGF D+DR++ S PS M S + SS TG
Sbjct: 204 MQALPICSRLLNQEILRVSG---MLSNQGFGDFDRLRHRS--PSPMASSNLMSSVTGTGL 368
Query: 58 --WNSLSHE------GLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRG 109
WNSL E G+ +DWQ APA +S+ VK++LRL+IP D +P FNFVGRLLGPRG
Sbjct: 369 GGWNSLQQERLRGTPGMAMDWQVAPASPSSYTVKRILRLEIPVDTYPNFNFVGRLLGPRG 548
Query: 110 NSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLR 169
NSLKRVEA+TGCRV+IRGKGSIKD DKEE LRGRPG+EHLNE LHILIEA+LP NVVDLR
Sbjct: 549 NSLKRVEASTGCRVYIRGKGSIKDPDKEEKLRGRPGYEHLNEQLHILIEADLPANVVDLR 728
Query: 170 LRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNEMIKRA 229
LRQAQEIIEELLKPV+ES+D KRQQLRELAMLNS+FREESP SGS+SPF S+ M KRA
Sbjct: 729 LRQAQEIIEELLKPVEESEDYIKRQQLRELAMLNSNFREESPGPSGSVSPFNSSGM-KRA 905
Query: 230 KT 231
KT
Sbjct: 906 KT 911
>TC215132 similar to UP|Q75GR5 (Q75GR5) Expressed protein, partial (55%)
Length = 753
Score = 284 bits (726), Expect = 3e-77
Identities = 139/169 (82%), Positives = 157/169 (92%)
Frame = +3
Query: 64 EGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRV 123
+GLN+DWQ +P + +S IVKK+LRLDIP D++P FNFVGRLLGPRGNSLKRVEATTGCRV
Sbjct: 33 QGLNMDWQTSPNVPSSPIVKKILRLDIPKDSYPNFNFVGRLLGPRGNSLKRVEATTGCRV 212
Query: 124 FIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKP 183
FIRGKGSIKD DKEE+LRGRPG+EHLN+PLHI+IEAELP +V D+RL QAQEII+ELLKP
Sbjct: 213 FIRGKGSIKDLDKEEMLRGRPGYEHLNDPLHIIIEAELPTSVADVRLMQAQEIIQELLKP 392
Query: 184 VDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNEMIKRAKTD 232
VDESQD+YKRQQLRELAMLNS+FREESPQLSGS+SPFTSNE IKR KTD
Sbjct: 393 VDESQDLYKRQQLRELAMLNSNFREESPQLSGSVSPFTSNE-IKRVKTD 536
>TC219760 weakly similar to UP|O88972 (O88972) KH domain RNA binding protein
QKI-7B, partial (18%)
Length = 957
Score = 264 bits (674), Expect = 3e-71
Identities = 141/212 (66%), Positives = 156/212 (73%), Gaps = 43/212 (20%)
Frame = +1
Query: 64 EGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPT-------------------------- 97
+GLNVDWQ +P + +SHIVK+ LRLDI +D++P
Sbjct: 10 QGLNVDWQTSPGVPSSHIVKRTLRLDIANDSYPNVRRYVEHFNAFCCILMKIYTMHCSNS 189
Query: 98 -----------------FNFVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELL 140
FN VGRLLGPRGNSLKRVEATTGCRVFIRGKGSIK+ DKEELL
Sbjct: 190 FMVLY*ALRH*A*LKFQFNLVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKELDKEELL 369
Query: 141 RGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQQLRELA 200
RGRPG+EHLNEPLH+LIEAELPVNVVD+RLRQAQEIIEELLKP+DESQD+YKRQQLRELA
Sbjct: 370 RGRPGYEHLNEPLHVLIEAELPVNVVDIRLRQAQEIIEELLKPMDESQDLYKRQQLRELA 549
Query: 201 MLNSSFREESPQLSGSLSPFTSNEMIKRAKTD 232
MLNS+FREESPQLS S S F SNEM KRAKTD
Sbjct: 550 MLNSNFREESPQLSASPSTFNSNEM-KRAKTD 642
>TC204004 similar to UP|Q8W574 (Q8W574) At1g09660/F21M12_5, partial (62%)
Length = 1255
Score = 204 bits (519), Expect = 3e-53
Identities = 120/235 (51%), Positives = 153/235 (65%), Gaps = 4/235 (1%)
Frame = +1
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 60
+QVLP T+LL QEI R S G G ++ T P +D GW +
Sbjct: 232 LQVLPQSTKLLTQEIRRMSVGGG----GGGGGFNHEPAADTPPPYFRPMD----LEGW-A 384
Query: 61 LSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTG 120
+ + + QR A + +VK+++RLD+P D P +NFVGR+LGPRGNSLKRVEA T
Sbjct: 385 IEVQQDKPNPQRMMAWP-APVVKRVIRLDVPVDKFPNYNFVGRILGPRGNSLKRVEAMTE 561
Query: 121 CRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEEL 180
CRV+IRG GS+KD KEE L+ +PG+EHL EPLH+L+EAE P ++++ RL A I+E L
Sbjct: 562 CRVYIRGCGSVKDSIKEEKLKEKPGYEHLKEPLHVLVEAEFPEDIINARLDHAVAILENL 741
Query: 181 LKPVDESQDIYKRQQLRELAMLNSSFREE----SPQLSGSLSPFTSNEMIKRAKT 231
LKPVDES D YK+QQLRELAMLN + REE SP +S S+SPF S M KRAKT
Sbjct: 742 LKPVDESLDHYKKQQLRELAMLNGTLREESPSMSPSMSPSMSPFNSTGM-KRAKT 903
>TC217026 weakly similar to UP|O88972 (O88972) KH domain RNA binding protein
QKI-7B, partial (19%)
Length = 696
Score = 203 bits (516), Expect = 6e-53
Identities = 105/163 (64%), Positives = 123/163 (75%), Gaps = 9/163 (5%)
Frame = +2
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTG--- 57
MQVLP+C+RLLNQEILR SG ++ NQGF D+DR++ S P +L + S TG
Sbjct: 215 MQVLPICSRLLNQEILRVSG---MLSNQGFGDFDRLRHRSPSPMASSNLMSNVSGTGLGG 385
Query: 58 WNSLSHE------GLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNS 111
WNSL E G+ +DWQ APA +S VK++LRL+IP D +P FNFVGRLLGPRGNS
Sbjct: 386 WNSLQQERLCGAPGMTMDWQSAPASPSSFTVKRILRLEIPVDTYPNFNFVGRLLGPRGNS 565
Query: 112 LKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLH 154
LKRVEATTGCRV+IRGKGSIKD DKEE LRGRPG+EHLNEPLH
Sbjct: 566 LKRVEATTGCRVYIRGKGSIKDPDKEEKLRGRPGYEHLNEPLH 694
>TC217027 weakly similar to UP|O88972 (O88972) KH domain RNA binding protein
QKI-7B, partial (6%)
Length = 921
Score = 154 bits (390), Expect = 3e-38
Identities = 81/95 (85%), Positives = 86/95 (90%)
Frame = +2
Query: 137 EELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQQL 196
EE LRGRPG+EHLNEPLHILIEAELP NVVD+RLRQAQEIIEELLKPVDESQD KRQQL
Sbjct: 2 EEKLRGRPGYEHLNEPLHILIEAELPANVVDIRLRQAQEIIEELLKPVDESQDYIKRQQL 181
Query: 197 RELAMLNSSFREESPQLSGSLSPFTSNEMIKRAKT 231
RELAMLNS+FREESP SGS+SPF S+ M KRAKT
Sbjct: 182 RELAMLNSNFREESPGPSGSVSPFNSSGM-KRAKT 283
>TC203784
Length = 787
Score = 105 bits (261), Expect = 2e-23
Identities = 55/70 (78%), Positives = 62/70 (88%)
Frame = +2
Query: 162 PVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFT 221
P N+VD+RLRQAQEIIEELLKPV+ES+D KRQQLRELAMLNS+FREESP SGS+SPF
Sbjct: 2 PANIVDIRLRQAQEIIEELLKPVEESEDYIKRQQLRELAMLNSNFREESPGPSGSVSPFN 181
Query: 222 SNEMIKRAKT 231
S+ M KRAKT
Sbjct: 182 SSGM-KRAKT 208
>TC209308 similar to UP|Q75GR5 (Q75GR5) Expressed protein, partial (14%)
Length = 943
Score = 87.4 bits (215), Expect = 5e-18
Identities = 43/64 (67%), Positives = 53/64 (82%)
Frame = +2
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 60
MQVLPLCTRL+NQEILR +GK+ L+QNQGFSD+DR++F S M S ++T +FTGWNS
Sbjct: 392 MQVLPLCTRLINQEILRVTGKNELLQNQGFSDFDRMRF--INLSHMASPNSTPNFTGWNS 565
Query: 61 LSHE 64
LSHE
Sbjct: 566 LSHE 577
>BG237608 homologue to PIR|T01782|T017 GDP dissociation inhibitor - common
tobacco, partial (13%)
Length = 432
Score = 72.8 bits (177), Expect = 1e-13
Identities = 32/43 (74%), Positives = 39/43 (90%)
Frame = +2
Query: 128 KGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRL 170
KGSIKD DKE +LRG+PG+EHLN+PLHI+IEAELP +V D+RL
Sbjct: 302 KGSIKDLDKE*MLRGKPGYEHLNDPLHIIIEAELPTSVADVRL 430
>TC213763 similar to UP|Q75GR5 (Q75GR5) Expressed protein, partial (13%)
Length = 616
Score = 70.1 bits (170), Expect = 8e-13
Identities = 33/36 (91%), Positives = 34/36 (93%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRV 36
MQVLPLCTRLLNQEILR SGK+GLMQNQG SDYDRV
Sbjct: 507 MQVLPLCTRLLNQEILRVSGKNGLMQNQGLSDYDRV 614
>BE805327 similar to GP|20161243|dbj P0408G07.16 {Oryza sativa (japonica
cultivar-group)}, partial (17%)
Length = 145
Score = 60.1 bits (144), Expect = 8e-10
Identities = 26/47 (55%), Positives = 34/47 (72%)
Frame = +2
Query: 81 IVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFIRG 127
+VK+++ LD+P D P +N+VGR+LGP GN LKRVEA T C I G
Sbjct: 5 VVKRVIMLDVPVDKFPNYNYVGRILGPGGNFLKRVEAMTECMAHITG 145
>BE059640
Length = 165
Score = 53.5 bits (127), Expect = 8e-08
Identities = 27/32 (84%), Positives = 28/32 (87%)
Frame = +1
Query: 185 DESQDIYKRQQLRELAMLNSSFREESPQLSGS 216
DESQD KRQQLRELAMLNS+FREESP SGS
Sbjct: 70 DESQDYIKRQQLRELAMLNSNFREESPGPSGS 165
>TC204000 homologue to UP|Q84XC3 (Q84XC3) Ubiquitin/ribosomal fusion protein,
partial (56%)
Length = 1061
Score = 47.8 bits (112), Expect = 4e-06
Identities = 28/47 (59%), Positives = 33/47 (69%), Gaps = 4/47 (8%)
Frame = +2
Query: 189 DIYKRQQLRELAMLNSSFREE----SPQLSGSLSPFTSNEMIKRAKT 231
D YK+Q LRELAMLN + REE SP +S S+SPF S M KRA+T
Sbjct: 596 DHYKKQPLRELAMLNGTLREESPSMSPSMSPSMSPFNSTGM-KRAQT 733
>TC230469 similar to GB|BAB09296.1|9758634|AB011476 RNA-binding protein-like
{Arabidopsis thaliana;} , partial (32%)
Length = 476
Score = 41.2 bits (95), Expect = 4e-04
Identities = 38/112 (33%), Positives = 50/112 (43%), Gaps = 21/112 (18%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRAS---------GKSGL----------MQNQGFSDYD--RVQFG 39
M VLP C RLLNQEILR + G+SGL + + G +D + +F
Sbjct: 186 MAVLPHCFRLLNQEILRVTTLMGNASVLGQSGLEHASPLATGGIFSNGGADVNGWASRFQ 365
Query: 40 STKPSLMPSLDTTSSFTGWNSLSHEGLNVDWQRAPAISNSHIVKKMLRLDIP 91
S +PSL+ S T S W S+ IVKK +R+DIP
Sbjct: 366 SERPSLLQSSSTQS----------------WLSPQGSSSGIIVKKTVRVDIP 473
>AW757164
Length = 388
Score = 40.8 bits (94), Expect = 5e-04
Identities = 26/68 (38%), Positives = 38/68 (55%), Gaps = 5/68 (7%)
Frame = +2
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTT-----SSF 55
MQVL +C RLL Q++ R SG++ +Q D+D+++ S P+ M S D +
Sbjct: 128 MQVLSICMRLLRQKMWRV---SGMLDHQRCGDFDKLRHRS--PNTMAS*DLMYYVCWTRL 292
Query: 56 TGWNSLSH 63
GWNSL H
Sbjct: 293 GGWNSLHH 316
>AI938973
Length = 408
Score = 30.8 bits (68), Expect = 0.55
Identities = 16/30 (53%), Positives = 20/30 (66%)
Frame = +2
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGF 30
+QVL LL EILR +GK+ +QNQGF
Sbjct: 317 VQVLIFTIILLLSEILRVTGKNESLQNQGF 406
>BF068195
Length = 365
Score = 28.9 bits (63), Expect = 2.1
Identities = 14/23 (60%), Positives = 15/23 (64%)
Frame = +1
Query: 1 MQVLPLCTRLLNQEILRASGKSG 23
M VLP C RL NQEILR + G
Sbjct: 199 MAVLPHCFRLFNQEILRVTTLMG 267
>TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division protein ftsH
homolog, chloroplast precursor , partial (88%)
Length = 2059
Score = 26.9 bits (58), Expect = 7.9
Identities = 26/100 (26%), Positives = 46/100 (46%), Gaps = 2/100 (2%)
Frame = +1
Query: 122 RVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELL 181
+V RGK KD D E++ R PGF + +++ EA + DL+ EI + L
Sbjct: 1063 QVHSRGKALAKDVDFEKIARRTPGFTGA-DLQNLMNEAAILAARRDLKEISKDEISDALE 1239
Query: 182 KPV--DESQDIYKRQQLRELAMLNSSFREESPQLSGSLSP 219
+ + E ++ + ++L ++ E L G+L P
Sbjct: 1240 RIIAGPEKKNAVVSDEKKKLV----AYHEAGHALVGALMP 1347
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,927,599
Number of Sequences: 63676
Number of extensions: 113503
Number of successful extensions: 562
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 554
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 555
length of query: 232
length of database: 12,639,632
effective HSP length: 94
effective length of query: 138
effective length of database: 6,654,088
effective search space: 918264144
effective search space used: 918264144
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149600.13


