

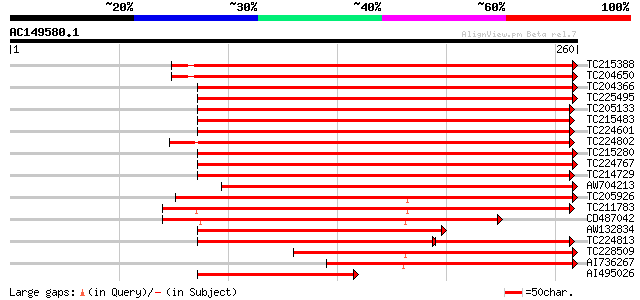
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149580.1 + phase: 0 /pseudo
(260 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215388 homologue to UP|Q42780 (Q42780) Lipoxygenase , complete 276 9e-75
TC204650 UP|Q43446 (Q43446) Lipoxygenase L-5 , complete 276 9e-75
TC204366 UP|LOX4_SOYBN (P38417) Lipoxygenase-4 (L-4) (VSP94) , ... 273 7e-74
TC225495 PDB|1LNH.0|1942405|1LNH Lipoxygenase-3(Soybean) Non-Hem... 265 1e-71
TC205133 UP|LOX2_SOYBN (P09439) Seed lipoxygenase-2 (L-2) , com... 263 6e-71
TC215483 UP|LOX1_SOYBN (P08170) Seed lipoxygenase-1 (L-1) , com... 262 1e-70
TC224601 homologue to UP|O24320 (O24320) Lipoxygenase , partial... 258 2e-69
TC224802 similar to UP|O24320 (O24320) Lipoxygenase , partial (... 254 4e-68
TC215280 UP|Q43440 (Q43440) Lipoxygenase , complete 253 6e-68
TC224767 lipoxygenase [Glycine max] 250 4e-67
TC214729 UP|LOXX_SOYBN (P24095) Seed lipoxygenase , complete 250 5e-67
AW704213 similar to PIR|T06596|T06 lipoxygenase (EC 1.13.11.12) ... 215 1e-56
TC205926 similar to UP|Q6X5R6 (Q6X5R6) Lipoxygenase (Fragment), ... 206 9e-54
TC211783 weakly similar to UP|Q6X5R5 (Q6X5R5) Lipoxygenase, part... 191 2e-49
CD487042 similar to GP|18461098|db lipoxygenase {Citrus jambhiri... 173 8e-44
AW132834 similar to GP|7340729|emb lipoxygenase-9 {Cucumis sativ... 164 3e-41
TC224813 homologue to UP|LOXX_SOYBN (P24095) Seed lipoxygenase ... 157 6e-39
TC228509 similar to UP|Q8W418 (Q8W418) Lipoxygenase, partial (35%) 135 2e-32
AI736267 129 2e-30
AI495026 similar to PIR|S18612|S18 lipoxygenase (EC 1.13.11.12) ... 118 2e-27
>TC215388 homologue to UP|Q42780 (Q42780) Lipoxygenase , complete
Length = 2995
Score = 276 bits (705), Expect = 9e-75
Identities = 130/186 (69%), Positives = 153/186 (81%)
Frame = +3
Query: 75 PNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHK 134
P+S A + WLLAKAY VVNDSC HQLVSHWL THAVVEPFVIATNRHLSV+HPI+K
Sbjct: 1527 PSSEGVEA--YIWLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFVIATNRHLSVVHPIYK 1700
Query: 135 LLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLK 194
LL PHYR TM IN+ +R L+NA GI+E TFL+ +Y +EMSAV+YKDWVF ++ LP DL
Sbjct: 1701 LLFPHYRDTMNINSLARKSLVNADGIIEKTFLWGRYSLEMSAVIYKDWVFTDQALPNDLV 1880
Query: 195 NRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELK 254
RG+AV D S+PHG+RLLIEDYPYASDGLEIW AIKSWV EYV+FYY+SD ++ D EL+
Sbjct: 1881 KRGVAVKDPSAPHGVRLLIEDYPYASDGLEIWDAIKSWVHEYVSFYYKSDAAIQQDPELQ 2060
Query: 255 AFWKEL 260
A+WKEL
Sbjct: 2061 AWWKEL 2078
>TC204650 UP|Q43446 (Q43446) Lipoxygenase L-5 , complete
Length = 2946
Score = 276 bits (705), Expect = 9e-75
Identities = 127/186 (68%), Positives = 156/186 (83%)
Frame = +3
Query: 75 PNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHK 134
P+S A + WLLAKAY VVND+C HQ++SHWL THAVVEPFVIATNRHLSV+HPI+K
Sbjct: 1470 PSSEGVEA--YIWLLAKAYVVVNDACYHQIISHWLNTHAVVEPFVIATNRHLSVVHPIYK 1643
Query: 135 LLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLK 194
LL PHYR TM IN+ +R L+NA GI+E TFL+ +Y +EMSAV+YKDWVF ++ LP DL
Sbjct: 1644 LLFPHYRDTMNINSLARKSLVNADGIIEKTFLWGRYSLEMSAVIYKDWVFTDQALPNDLV 1823
Query: 195 NRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELK 254
RG+AV D S+PHG+RLLIEDYPYASDGLEIW AIKSWV+EYV+FYY+SD++++ D EL+
Sbjct: 1824 KRGVAVKDPSAPHGVRLLIEDYPYASDGLEIWDAIKSWVEEYVSFYYKSDEELQKDPELQ 2003
Query: 255 AFWKEL 260
A+WKEL
Sbjct: 2004 AWWKEL 2021
>TC204366 UP|LOX4_SOYBN (P38417) Lipoxygenase-4 (L-4) (VSP94) , complete
Length = 3220
Score = 273 bits (697), Expect = 7e-74
Identities = 123/174 (70%), Positives = 149/174 (84%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVND+C HQ++SHWL THA+VEPFVIATNR LSV+HPI+KLL PHYR TM I
Sbjct: 1541 WLLAKAYVVVNDACYHQIISHWLSTHAIVEPFVIATNRQLSVVHPIYKLLFPHYRDTMNI 1720
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+NA GI+E TFL+ +Y MEMSAV+YKDWVF ++ LP DL RG+AV D S+P
Sbjct: 1721 NSLARKALVNADGIIEKTFLWGRYSMEMSAVIYKDWVFTDQALPNDLVKRGVAVKDPSAP 1900
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIW AIKSWV EYV+FYY+SD++++ D EL+A+WKEL
Sbjct: 1901 HGVRLLIEDYPYASDGLEIWDAIKSWVQEYVSFYYKSDEELQKDPELQAWWKEL 2062
>TC225495 PDB|1LNH.0|1942405|1LNH Lipoxygenase-3(Soybean) Non-Heme Fe(Ii)
Metalloprotein. {Glycine max;} , complete
Length = 2826
Score = 265 bits (678), Expect = 1e-71
Identities = 122/174 (70%), Positives = 144/174 (82%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPI+KLL PHYR TM I
Sbjct: 1540 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIYKLLHPHYRDTMNI 1719
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R L+N GG++E TFL+ +Y +EMSAVVYKDWVF ++ LP DL RGMA++D S P
Sbjct: 1720 NGLARLSLVNDGGVIEQTFLWGRYSVEMSAVVYKDWVFTDQALPADLIKRGMAIEDPSCP 1899
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL+IEDYPYA DGLEIW AIK+WV EYV YY+SD +++D EL+A WKEL
Sbjct: 1900 HGIRLVIEDYPYAVDGLEIWDAIKTWVHEYVFLYYKSDDTLREDPELQACWKEL 2061
>TC205133 UP|LOX2_SOYBN (P09439) Seed lipoxygenase-2 (L-2) , complete
Length = 2936
Score = 263 bits (672), Expect = 6e-71
Identities = 121/173 (69%), Positives = 141/173 (80%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQL+SHWL THAV+EPF+IATNRHLS LHPI+KLL PHYR TM I
Sbjct: 1585 WLLAKAYVVVNDSCYHQLMSHWLNTHAVIEPFIIATNRHLSALHPIYKLLTPHYRDTMNI 1764
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINA GI+E +FL K+ +EMS+ VYK+WVF ++ LP DL RG+A+ D S+P
Sbjct: 1765 NALARQSLINADGIIEKSFLPSKHSVEMSSAVYKNWVFTDQALPADLIKRGVAIKDPSAP 1944
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HGLRLLIEDYPYA DGLEIWAAIK+WV EYV+ YY D DVK D EL+ +WKE
Sbjct: 1945 HGLRLLIEDYPYAVDGLEIWAAIKTWVQEYVSLYYARDDDVKPDSELQQWWKE 2103
>TC215483 UP|LOX1_SOYBN (P08170) Seed lipoxygenase-1 (L-1) , complete
Length = 2953
Score = 262 bits (670), Expect = 1e-70
Identities = 120/173 (69%), Positives = 142/173 (81%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQL+SHWL THA +EPFVIAT+RHLSVLHPI+KLL PHYR M I
Sbjct: 1522 WLLAKAYVIVNDSCYHQLMSHWLNTHAAMEPFVIATHRHLSVLHPIYKLLTPHYRNNMNI 1701
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINA GI+E+TFL KY +EMS+ VYK+WVF ++ LP DL RG+A+ D S+P
Sbjct: 1702 NALARQSLINANGIIETTFLPSKYSVEMSSAVYKNWVFTDQALPADLIKRGVAIKDPSTP 1881
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HG+RLLIEDYPYA+DGLEIWAAIK+WV EYV YY D DVK+D EL+ +WKE
Sbjct: 1882 HGVRLLIEDYPYAADGLEIWAAIKTWVQEYVPLYYARDDDVKNDSELQHWWKE 2040
>TC224601 homologue to UP|O24320 (O24320) Lipoxygenase , partial (52%)
Length = 1668
Score = 258 bits (658), Expect = 2e-69
Identities = 118/173 (68%), Positives = 143/173 (82%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WL+AKAY VVNDSC HQL+SHWL THAV+EPFVIATNRHLSVLHPI+KLL+PHYR TM I
Sbjct: 260 WLIAKAYVVVNDSCYHQLMSHWLNTHAVIEPFVIATNRHLSVLHPIYKLLLPHYRDTMNI 439
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINAGGI+E +FL + +EMS+ VYK WVF ++ LP DL RGMAV+D SSP
Sbjct: 440 NGLARQSLINAGGIIEQSFLPGPFAVEMSSAVYKGWVFTDQALPADLIKRGMAVEDPSSP 619
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
+GLRL+I+DYPYA DGLEIW+AI++WV +YV+ YY +D VK D EL+A+WKE
Sbjct: 620 YGLRLVIDDYPYAVDGLEIWSAIQTWVKDYVSLYYATDDAVKKDSELQAWWKE 778
>TC224802 similar to UP|O24320 (O24320) Lipoxygenase , partial (73%)
Length = 2213
Score = 254 bits (648), Expect = 4e-68
Identities = 118/186 (63%), Positives = 143/186 (76%)
Frame = +2
Query: 74 PPNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIH 133
PP + GT WL+AKAY VND+ HQL+SHWL THA +EPFVIATNRHLSVLHPIH
Sbjct: 803 PPAAVNSAEGTI-WLIAKAYVAVNDTGYHQLISHWLNTHATIEPFVIATNRHLSVLHPIH 979
Query: 134 KLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDL 193
KLL+PHYR TM INA +R LINA G++E +FL KY +EMS+ VYK WVF ++ LP DL
Sbjct: 980 KLLLPHYRDTMNINALARQSLINADGVIERSFLPGKYSLEMSSAVYKSWVFTDQALPADL 1159
Query: 194 KNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEEL 253
RGMA++D +PHGLRL+IEDYPYA DGLEIW AI++WV YV+ YY +D +K D EL
Sbjct: 1160IKRGMAIEDPCAPHGLRLVIEDYPYAVDGLEIWDAIQTWVKNYVSLYYPTDDAIKKDSEL 1339
Query: 254 KAFWKE 259
+A+WKE
Sbjct: 1340QAWWKE 1357
>TC215280 UP|Q43440 (Q43440) Lipoxygenase , complete
Length = 2894
Score = 253 bits (646), Expect = 6e-68
Identities = 116/174 (66%), Positives = 143/174 (81%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQL+SHWL THAV+EPF IATNRHLSVLHPI+KLL PHY+ T+ I
Sbjct: 1503 WLLAKAHVIVNDSGYHQLISHWLNTHAVMEPFAIATNRHLSVLHPIYKLLYPHYKDTINI 1682
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINAGGI+E TFL KY +EMS+VVYK+WVF ++ LP DL RG+AV+D S+P
Sbjct: 1683 NGLARQSLINAGGIIEQTFLPGKYSIEMSSVVYKNWVFTDQALPADLVKRGLAVEDPSAP 1862
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HGLRL+IEDYPYA DGLEIW AIK+WV EYV+ YY ++ ++ D EL+A+WKE+
Sbjct: 1863 HGLRLVIEDYPYAVDGLEIWDAIKTWVHEYVSVYYPTNAAIQQDTELQAWWKEV 2024
>TC224767 lipoxygenase [Glycine max]
Length = 1729
Score = 250 bits (639), Expect = 4e-67
Identities = 114/174 (65%), Positives = 141/174 (80%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDSC HQL+SHWL THAV+EPFVIATNR+LS+LHPI+KLL PHYR TM I
Sbjct: 538 WLLAKAHVIVNDSCYHQLISHWLNTHAVIEPFVIATNRNLSILHPIYKLLFPHYRDTMNI 717
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINA G +E TFL KY +E+S+ YK+WVF ++ LP DL RGMA++D S P
Sbjct: 718 NALARQSLINADGFIEKTFLGGKYAVEISSSGYKNWVFLDQALPADLIKRGMAIEDSSCP 897
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+GLRL+IEDYPYA DGLEIW AIK+WV EYV+ YY ++ +K D EL+A+WKE+
Sbjct: 898 NGLRLVIEDYPYAVDGLEIWDAIKTWVQEYVSLYYATNDAIKKDHELQAWWKEV 1059
>TC214729 UP|LOXX_SOYBN (P24095) Seed lipoxygenase , complete
Length = 2891
Score = 250 bits (638), Expect = 5e-67
Identities = 116/173 (67%), Positives = 140/173 (80%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQLVSHWL THAV+EPF IATNRHLSVLHPI+KLL PHYR T+ I
Sbjct: 1589 WLLAKAHVIVNDSGYHQLVSHWLNTHAVMEPFAIATNRHLSVLHPIYKLLYPHYRDTINI 1768
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINA GI+E +FL KY +EMS+ VYK+WVF ++ LP DL RG+A++D S+P
Sbjct: 1769 NGLARQSLINADGIIEKSFLPGKYSIEMSSSVYKNWVFTDQALPADLVKRGLAIEDPSAP 1948
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HGLRL+IEDYPYA DGLEIW AIK+WV EYV+ YY +D V+ D EL+A+WKE
Sbjct: 1949 HGLRLVIEDYPYAVDGLEIWDAIKTWVHEYVSLYYPTDAAVQQDTELQAWWKE 2107
>AW704213 similar to PIR|T06596|T06 lipoxygenase (EC 1.13.11.12) 7 - soybean,
partial (22%)
Length = 589
Score = 215 bits (548), Expect = 1e-56
Identities = 104/163 (63%), Positives = 124/163 (75%)
Frame = +2
Query: 98 DSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTINARSRNILINA 157
+S HQL +HWL THAVVEPFVIATNRH++ HPI KL PHYR TM IN+ +R L+ A
Sbjct: 2 ESSYHQLGNHWLNTHAVVEPFVIATNRHMNAAHPIDKL*GPHYRDTMNINSLARKCLVKA 181
Query: 158 GGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYP 217
GI++ TFL +Y +EMSAV+YKDWVF ++ LP DL RG+A D S+PHG+RL IEDYP
Sbjct: 182 DGIIQKTFLRGRYSLEMSAVMYKDWVFTDQALPNDLVKRGVADKDPSAPHGVRLSIEDYP 361
Query: 218 YASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
YASDGLEIW AIKS V EYV+FYY+SD D EL+A WKEL
Sbjct: 362 YASDGLEIWDAIKS*VHEYVSFYYKSDASR*QDPELRA*WKEL 490
>TC205926 similar to UP|Q6X5R6 (Q6X5R6) Lipoxygenase (Fragment), partial
(57%)
Length = 1965
Score = 206 bits (524), Expect = 9e-54
Identities = 95/185 (51%), Positives = 133/185 (71%), Gaps = 1/185 (0%)
Frame = +2
Query: 77 STAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLL 136
S ++ + W LAKA+ + +D+ H+L++HWL+THAV+EPFV+ATNR LSV+HPI+KLL
Sbjct: 695 SISHSTNLWLWRLAKAHVLAHDAGVHELINHWLRTHAVMEPFVVATNRQLSVMHPIYKLL 874
Query: 137 VPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD-WVFAEEGLPTDLKN 195
PH T+ IN+ +R ILIN GI+E +F KY ME+S+ Y W F + LP DL +
Sbjct: 875 HPHLTYTLAINSLAREILINGNGIIEKSFSPNKYSMELSSAAYDQLWRFDLQALPNDLID 1054
Query: 196 RGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKA 255
RG+AV D ++PHGL+L IEDYP+A+DGL IW AIK W+ EYVN YY + ++ D+EL+
Sbjct: 1055RGIAVVDPNAPHGLKLTIEDYPFANDGLLIWDAIKQWITEYVNHYYPTPSIIESDQELQP 1234
Query: 256 FWKEL 260
+W E+
Sbjct: 1235WWTEI 1249
>TC211783 weakly similar to UP|Q6X5R5 (Q6X5R5) Lipoxygenase, partial (44%)
Length = 1338
Score = 191 bits (486), Expect = 2e-49
Identities = 97/198 (48%), Positives = 128/198 (63%), Gaps = 9/198 (4%)
Frame = +2
Query: 71 PPGPPNSTAYHAGT--------FNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIAT 122
P G PN+++ T + W L KA+ ND+ H LV HWL+ HA +EP +IAT
Sbjct: 74 PTGNPNTSSKQVLTPPKDATSKWLWQLGKAHVCSNDAGVHTLVHHWLRIHACMEPLIIAT 253
Query: 123 NRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD- 181
+R LSV+HPI KLL PH R T+ NA +R LINA G +E+ +YCM+ S+ YKD
Sbjct: 254 HRQLSVMHPIFKLLHPHMRYTLKTNAIARQTLINAEGTIETDHTPGRYCMQFSSAAYKDW 433
Query: 182 WVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYY 241
W F EG P DL RG+AV D + PHG+RLLIEDYPYA+DGL IW++IK V YVN YY
Sbjct: 434 WRFDMEGFPADLIRRGLAVPDATQPHGIRLLIEDYPYAADGLLIWSSIKKLVRTYVNHYY 613
Query: 242 ESDKDVKDDEELKAFWKE 259
++ V D EL+++++E
Sbjct: 614 KNSNAVSSDNELQSWYRE 667
>CD487042 similar to GP|18461098|db lipoxygenase {Citrus jambhiri}, partial
(22%)
Length = 638
Score = 173 bits (438), Expect = 8e-44
Identities = 88/167 (52%), Positives = 111/167 (65%), Gaps = 11/167 (6%)
Frame = +1
Query: 71 PPGPPNSTAYHAGTFN----------WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVI 120
PP N+T F W AKA+ + +DS HQLVSHWL+TH V EP+VI
Sbjct: 133 PPSSSNNTGQWKQVFTPSWHSTSVRLWRFAKAHVLAHDSGYHQLVSHWLRTHCVTEPYVI 312
Query: 121 ATNRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYK 180
ATNR LS LHPI+KLL PH+R TM INA +R LINA G +ES+F KY +E+S+ Y
Sbjct: 313 ATNRQLSELHPIYKLLHPHFRYTMEINAIAREALINADGTIESSFAPGKYSIEISSAAYA 492
Query: 181 -DWVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIW 226
+W F ++ LP DL +RGMAV D SPHGL+L I+DYP+A+DGL +W
Sbjct: 493 LEWRFDKQALPADLVSRGMAVKDPFSPHGLKLTIQDYPFANDGLLLW 633
>AW132834 similar to GP|7340729|emb lipoxygenase-9 {Cucumis sativus}, partial
(15%)
Length = 425
Score = 164 bits (416), Expect = 3e-41
Identities = 77/114 (67%), Positives = 90/114 (78%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAYA VNDS HQLVSHWL THAV+EPF+I TNR LS+LHPIHKLL PH+R TM I
Sbjct: 81 WQLAKAYAAVNDSGYHQLVSHWLYTHAVIEPFIITTNRQLSILHPIHKLLKPHFRDTMHI 260
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAV 200
NA +R+ LINAGG++E T K+ +EMS+V+YK WVF E+ LP DL RGMAV
Sbjct: 261 NALARHTLINAGGVLEKTVFPGKFALEMSSVIYKSWVFTEQALPADLLKRGMAV 422
>TC224813 homologue to UP|LOXX_SOYBN (P24095) Seed lipoxygenase , partial
(34%)
Length = 1200
Score = 157 bits (396), Expect = 6e-39
Identities = 74/110 (67%), Positives = 87/110 (78%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQLVSHWL THAV+EPF IATNRHLSVLHPI+KLL PHYR T+ I
Sbjct: 147 WLLAKAHVIVNDSGYHQLVSHWLNTHAVMEPFAIATNRHLSVLHPIYKLLYPHYRDTINI 326
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNR 196
N +R LINA GI+E +FL KY +EMS+ VYK+WVF ++ LP DL R
Sbjct: 327 NGLARQSLINADGIIEKSFLPGKYSIEMSSSVYKNWVFTDQALPADLVKR 476
Score = 99.0 bits (245), Expect = 2e-21
Identities = 42/64 (65%), Positives = 54/64 (83%)
Frame = +1
Query: 196 RGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKA 255
RG+A++D S+PHGLRL+IEDYPYA DGLEIW AIK+WV EYV+ YY +D ++ D EL+A
Sbjct: 676 RGLAIEDPSAPHGLRLVIEDYPYAVDGLEIWDAIKTWVHEYVSLYYPTDAIIQQDAELQA 855
Query: 256 FWKE 259
+WKE
Sbjct: 856 WWKE 867
>TC228509 similar to UP|Q8W418 (Q8W418) Lipoxygenase, partial (35%)
Length = 1147
Score = 135 bits (340), Expect = 2e-32
Identities = 65/131 (49%), Positives = 90/131 (68%), Gaps = 1/131 (0%)
Frame = +3
Query: 131 PIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYK-DWVFAEEGL 189
P ++ P++R TM INA +R LINA G + S+F KY +E+S+ Y +W F ++ L
Sbjct: 9 PHKQIATPYFRYTMEINALAREALINADGTI*SSFAPGKYALEISSAAYALEWRFDKQAL 188
Query: 190 PTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKD 249
P DL RG+AV+D SPHGL+L I+DYP+A+DGL +W AIK WV +YVN YY V+
Sbjct: 189 PADLIRRGIAVEDPFSPHGLKLTIQDYPFANDGLLLWDAIKLWVTDYVNHYYPEPSLVES 368
Query: 250 DEELKAFWKEL 260
DEEL+A+W E+
Sbjct: 369 DEELQAWWTEI 401
>AI736267
Length = 422
Score = 129 bits (323), Expect = 2e-30
Identities = 59/116 (50%), Positives = 84/116 (71%), Gaps = 1/116 (0%)
Frame = +1
Query: 146 INARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNRGMAVDDDS 204
INA +R LIN GI+ES+F K+ + +S++ Y + W F + LP DL +RGMAV+D +
Sbjct: 4 INALAREALINGDGIIESSFSPGKHSILLSSIAYDQQWQFDLQSLPKDLISRGMAVEDPT 183
Query: 205 SPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+PHGL+L+IEDYPYA+DGL +W A+K+W EYVN YY D + D EL+A+W+E+
Sbjct: 184 APHGLKLIIEDYPYANDGLVLWDALKTWFTEYVNLYYADDGSIVSDTELQAWWEEI 351
>AI495026 similar to PIR|S18612|S18 lipoxygenase (EC 1.13.11.12) - soybean
(fragment), partial (21%)
Length = 404
Score = 118 bits (296), Expect = 2e-27
Identities = 54/74 (72%), Positives = 62/74 (82%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQL+SHWL THAV EPF+IATNR LSVLHPI+KLL PHYR T+ I
Sbjct: 182 WLLAKAHVIVNDSGYHQLMSHWLNTHAVTEPFIIATNRRLSVLHPIYKLLYPHYRDTINI 361
Query: 147 NARSRNILINAGGI 160
N +RN LINAGG+
Sbjct: 362 NGLARNALINAGGV 403
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,182,880
Number of Sequences: 63676
Number of extensions: 247395
Number of successful extensions: 4215
Number of sequences better than 10.0: 308
Number of HSP's better than 10.0 without gapping: 3500
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4031
length of query: 260
length of database: 12,639,632
effective HSP length: 95
effective length of query: 165
effective length of database: 6,590,412
effective search space: 1087417980
effective search space used: 1087417980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149580.1


