

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.2 + phase: 0
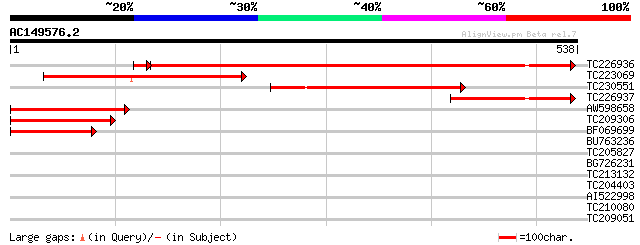
(538 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226936 similar to UP|Q71N54 (Q71N54) Diacylglycerol sterol acy... 688 0.0
TC223069 similar to UP|Q71LW1 (Q71LW1) Lecithin cholesterol acyl... 228 4e-60
TC230551 weakly similar to UP|Q71LW1 (Q71LW1) Lecithin cholester... 201 9e-52
TC226937 similar to UP|Q71N54 (Q71N54) Diacylglycerol sterol acy... 195 5e-50
AW598658 192 4e-49
TC209306 similar to UP|Q71N54 (Q71N54) Diacylglycerol sterol acy... 163 2e-40
BF069699 140 1e-33
BU763236 32 0.55
TC205827 32 0.71
BG726231 30 2.7
TC213132 similar to UP|ARFK_ARATH (Q9ZPY6) Auxin response factor... 29 4.6
TC204403 UP|SODC_SOYBN (Q7M1R5) Superoxide dismutase [Cu-Zn] , ... 29 4.6
AI522998 29 6.0
TC210080 weakly similar to UP|Q8VZH8 (Q8VZH8) AT4g30550/F17I23_1... 29 6.0
TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 28 7.9
>TC226936 similar to UP|Q71N54 (Q71N54) Diacylglycerol sterol
acyltransferase-like protein, partial (65%)
Length = 1586
Score = 688 bits (1775), Expect(2) = 0.0
Identities = 323/404 (79%), Positives = 364/404 (89%)
Frame = +3
Query: 134 GYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFM 193
G++EGKTLFGFGYDFRQSNRLQETMDR A KLE IYNAAGG KI++I+HSMGGLLVKCFM
Sbjct: 54 GFEEGKTLFGFGYDFRQSNRLQETMDRLAAKLESIYNAAGGNKINIITHSMGGLLVKCFM 233
Query: 194 TLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIEC 253
L SDIFEKYVKNW+AICAPFQGAPG NSTFLNGMSFVEGWEQNF+ISKWSMHQLLIEC
Sbjct: 234 CLQSDIFEKYVKNWVAICAPFQGAPGTINSTFLNGMSFVEGWEQNFYISKWSMHQLLIEC 413
Query: 254 PSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNH 313
PSIYELM CPN HW+H+P+LELWRER DGKSH++LESYPP DSIE+ KQAL+NN VN+
Sbjct: 414 PSIYELMGCPNSHWQHIPVLELWRERRDSDGKSHIVLESYPPCDSIEVLKQALLNNTVNY 593
Query: 314 EGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSD 373
G +LPLPFN I +WANKT EILSSAKLPS VKFYNIYGT+L TPHS+C+G+ DKPV+D
Sbjct: 594 NGVDLPLPFNLEILKWANKTWEILSSAKLPSQVKFYNIYGTSLDTPHSVCFGSGDKPVTD 773
Query: 374 LQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILCEPHLFRILKHWLK 433
LQ+LRY QA+YVCVDGDGTVP+ESAKADG NAE RVG+PGEH+ IL EPH+FR+LKHWLK
Sbjct: 774 LQQLRYFQAKYVCVDGDGTVPIESAKADGLNAEARVGVPGEHQRILREPHVFRLLKHWLK 953
Query: 434 AGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKDQDGQSNTGDNKMTL 493
AG+PDPFYNP+NDYVILPTAFEMERHKEKG+EVASLKEEWEIISK QD QS+T D +
Sbjct: 954 AGEPDPFYNPVNDYVILPTAFEMERHKEKGVEVASLKEEWEIISKVQDDQSSTADK---V 1124
Query: 494 SSISVSQEGANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVDA 537
SISVSQEGAN+S+SEAHATV VH D++GKQH++LNA+AVSVDA
Sbjct: 1125CSISVSQEGANQSYSEAHATVIVHPDSEGKQHVQLNALAVSVDA 1256
Score = 32.0 bits (71), Expect(2) = 0.0
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +1
Query: 118 EAVYYFHDMIVQMQKWG 134
++VYYFHDMIV+M+K G
Sbjct: 4 DSVYYFHDMIVEMRKXG 54
>TC223069 similar to UP|Q71LW1 (Q71LW1) Lecithin cholesterol
acyltransferase-like protein, partial (44%)
Length = 659
Score = 228 bits (582), Expect = 4e-60
Identities = 107/195 (54%), Positives = 140/195 (70%), Gaps = 3/195 (1%)
Frame = +2
Query: 33 DPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKS 92
DPVLLV G+GGSI+N+ + G RVWVR L A+ + + K+WS Y+P TG T TLD+KS
Sbjct: 74 DPVLLVSGMGGSIVNSKPKKFGFTTRVWVRLLLADVEFRNKIWSLYNPQTGYTETLDKKS 253
Query: 93 RIVVPEDRHGLHAIDVLDPDLV---IGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFR 149
IVVP+D HGL+AID+LDP I VY+FHDMI + GY +G TLFG+GYDFR
Sbjct: 254 EIVVPDDDHGLYAIDILDPSWFTKCIHLTEVYHFHDMIDMLVGCGYNKGTTLFGYGYDFR 433
Query: 150 QSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEKYVKNWIA 209
QSNR+ + M+ KLE + A+GG+K++LISHSMGG+++ CFM+L+ D+F KYV WI
Sbjct: 434 QSNRIGKVMEGLKSKLETAHKASGGRKVNLISHSMGGIMISCFMSLYRDVFTKYVNKWIC 613
Query: 210 ICAPFQGAPGCTNST 224
+ PFQGAPGC N +
Sbjct: 614 LACPFQGAPGCINDS 658
>TC230551 weakly similar to UP|Q71LW1 (Q71LW1) Lecithin cholesterol
acyltransferase-like protein, partial (41%)
Length = 893
Score = 201 bits (510), Expect = 9e-52
Identities = 93/185 (50%), Positives = 132/185 (71%)
Frame = +3
Query: 248 QLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALV 307
QLL+ECPSIYE++A P + WK P + +WR+ +DG +++ LESY P SI +F++AL
Sbjct: 3 QLLVECPSIYEMLANPYYEWKKQPEILVWRKHT-KDGDNNINLESYGPTQSISLFEEALR 179
Query: 308 NNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNA 367
+N+VN+ G+ + LPFN I +WA +TR+++++AKLP GV FYNIYGT+L TP +CYG+
Sbjct: 180 DNEVNYYGKTISLPFNFDILDWAVETRQLIANAKLPDGVCFYNIYGTSLDTPFDVCYGSE 359
Query: 368 DKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILCEPHLFRI 427
+ P+ DL E+ + Y VDGDGTVP ESAK DG A ERVG+ HRGIL + +F+
Sbjct: 360 NSPIEDLSEICHTMPLYSYVDGDGTVPSESAKGDGLEATERVGVAASHRGILRDETVFQH 539
Query: 428 LKHWL 432
++ WL
Sbjct: 540 IQKWL 554
>TC226937 similar to UP|Q71N54 (Q71N54) Diacylglycerol sterol
acyltransferase-like protein, partial (10%)
Length = 694
Score = 195 bits (495), Expect = 5e-50
Identities = 95/119 (79%), Positives = 107/119 (89%)
Frame = +1
Query: 419 LCEPHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISK 478
L EPH+FR+LKHWLKAG+PDPFYNP+NDYVILPTAFEMERHKEKG+EVASLKEEWEIISK
Sbjct: 1 LREPHVFRLLKHWLKAGEPDPFYNPVNDYVILPTAFEMERHKEKGVEVASLKEEWEIISK 180
Query: 479 DQDGQSNTGDNKMTLSSISVSQEGANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVDA 537
QD QS T D + SISVSQEGAN+S+SEAHATV VH DN+GKQH++LNA+AVSVDA
Sbjct: 181 VQDDQSCTADK---VCSISVSQEGANQSYSEAHATVIVHPDNEGKQHVQLNALAVSVDA 348
>AW598658
Length = 436
Score = 192 bits (487), Expect = 4e-49
Identities = 89/113 (78%), Positives = 105/113 (92%)
Frame = +3
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
M +LL ++L+S+ELWL+LIK PQPQ YVNPNLDPVLLVPGVGGS+L+AV+ES+GS+ERVW
Sbjct: 96 MAILLGEILQSLELWLKLIKNPQPQPYVNPNLDPVLLVPGVGGSMLHAVDESEGSRERVW 275
Query: 61 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDL 113
VRFL+AEY LKTKLWSRYDPSTGKT ++D SRI+VPEDRHGLHAID+LDPDL
Sbjct: 276 VRFLNAEYTLKTKLWSRYDPSTGKTESMDPNSRIMVPEDRHGLHAIDILDPDL 434
>TC209306 similar to UP|Q71N54 (Q71N54) Diacylglycerol sterol
acyltransferase-like protein, partial (7%)
Length = 462
Score = 163 bits (413), Expect = 2e-40
Identities = 76/100 (76%), Positives = 90/100 (90%)
Frame = +1
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
M +LL ++L+S+ELWL+LIK PQPQ YVNPNLDPVLLVPGVGGS+L+AV+E+DGS ERVW
Sbjct: 163 MAILLGEILQSLELWLKLIKNPQPQPYVNPNLDPVLLVPGVGGSMLHAVDETDGSHERVW 342
Query: 61 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDR 100
VRFL+AEY LKTKLWSRYDPSTGKT ++D S I+VPEDR
Sbjct: 343 VRFLNAEYTLKTKLWSRYDPSTGKTESMDPNSTIIVPEDR 462
>BF069699
Length = 406
Score = 140 bits (354), Expect = 1e-33
Identities = 65/82 (79%), Positives = 77/82 (93%)
Frame = +3
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
M +LL ++L+S+ELWL+LIK PQPQ YVNPNLDPVLLVPGVGGS+L+AV+ES+GS+ERVW
Sbjct: 114 MAILLGEILQSLELWLKLIKNPQPQPYVNPNLDPVLLVPGVGGSMLHAVDESEGSRERVW 293
Query: 61 VRFLSAEYKLKTKLWSRYDPST 82
VRFL+AEY LKTKLWSRYDPST
Sbjct: 294 VRFLNAEYTLKTKLWSRYDPST 359
>BU763236
Length = 420
Score = 32.3 bits (72), Expect = 0.55
Identities = 16/62 (25%), Positives = 31/62 (49%), Gaps = 2/62 (3%)
Frame = +3
Query: 231 FVEGWEQNFFISKWSMHQLLIECPSIY--ELMACPNFHWKHVPLLELWRERLHEDGKSHV 288
F+ W + ++ + +LI CP ++ L +C + H+ H+ W ER +SH+
Sbjct: 51 FLGPWLYHLLFNQLTKLLVLIYCPFLHLTPLNSCHHIHYSHLASFLEWLERCRLVAESHI 230
Query: 289 IL 290
+L
Sbjct: 231 LL 236
>TC205827
Length = 2695
Score = 32.0 bits (71), Expect = 0.71
Identities = 14/33 (42%), Positives = 19/33 (57%)
Frame = +3
Query: 201 EKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVE 233
E+ K W +C P+ G CT STFL ++VE
Sbjct: 492 EESFKAWGQVCVPYLG*ISCTKSTFLKVSTWVE 590
>BG726231
Length = 415
Score = 30.0 bits (66), Expect = 2.7
Identities = 36/135 (26%), Positives = 56/135 (40%)
Frame = +1
Query: 8 VLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAE 67
VL S L + KP P L G S ++ V S+G E + L AE
Sbjct: 1 VLMSPFAILGFVMKPLQLKGFAPTDSEKALTLGTVASEVSDVGASNGKDEAL---SLKAE 171
Query: 68 YKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMI 127
++ K S ++PS K LDQ SR + +D L +LD V+ ++ +I
Sbjct: 172 FRDK----SSHEPSRSKCTILDQFSRFL--KDMKEL----LLDKVFVVNVLGYIAYNFVI 321
Query: 128 VQMQKWGYQEGKTLF 142
WG + G +++
Sbjct: 322 GAYSYWGPKAGYSIY 366
>TC213132 similar to UP|ARFK_ARATH (Q9ZPY6) Auxin response factor 11, partial
(6%)
Length = 443
Score = 29.3 bits (64), Expect = 4.6
Identities = 12/32 (37%), Positives = 19/32 (58%)
Frame = -1
Query: 254 PSIYELMACPNFHWKHVPLLELWRERLHEDGK 285
P I ++AC N H H+PL + R++L G+
Sbjct: 362 PRIASIVACTNRHLPHLPLRKTERQKLKAGGR 267
>TC204403 UP|SODC_SOYBN (Q7M1R5) Superoxide dismutase [Cu-Zn] , complete
Length = 799
Score = 29.3 bits (64), Expect = 4.6
Identities = 16/48 (33%), Positives = 21/48 (43%), Gaps = 8/48 (16%)
Frame = +2
Query: 214 FQGAPGCTNSTFLNGMSFVEGWEQ-----NFFI---SKWSMHQLLIEC 253
F + GC T NG W+Q N+F+ KWS H+ I C
Sbjct: 38 FSKSKGCPEITLSNGEGCGSSWQQRGCHWNYFLHSGGKWSNHRNWISC 181
>AI522998
Length = 307
Score = 28.9 bits (63), Expect = 6.0
Identities = 11/29 (37%), Positives = 16/29 (54%)
Frame = +3
Query: 411 IPGEHRGILCEPHLFRILKHWLKAGDPDP 439
+P + I C PHLF +L W ++ P P
Sbjct: 114 LPPNTKWIACSPHLFFVLLEWSRSSIPWP 200
>TC210080 weakly similar to UP|Q8VZH8 (Q8VZH8) AT4g30550/F17I23_110, partial
(45%)
Length = 693
Score = 28.9 bits (63), Expect = 6.0
Identities = 15/41 (36%), Positives = 22/41 (53%)
Frame = -3
Query: 245 SMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGK 285
S H+ +IE I+ + P H HVP L L R+ HE+ +
Sbjct: 304 SDHKAIIETQVIF-IRKIPTRHLVHVPCLSLLRQHSHENSR 185
>TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (35%)
Length = 1017
Score = 28.5 bits (62), Expect = 7.9
Identities = 27/74 (36%), Positives = 38/74 (50%), Gaps = 2/74 (2%)
Frame = +2
Query: 431 WLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKDQDG--QSNTGD 488
+L+A DP LP+ F +ER KEKGL VAS + +++ + G S+ G
Sbjct: 182 YLEASKEDPLQ-------FLPSGF-LERTKEKGLVVASWAPQVQVLGHNSVGGFLSHCGW 337
Query: 489 NKMTLSSISVSQEG 502
N TL S+ QEG
Sbjct: 338 NS-TLESV---QEG 367
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,174,439
Number of Sequences: 63676
Number of extensions: 403402
Number of successful extensions: 1873
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1827
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1869
length of query: 538
length of database: 12,639,632
effective HSP length: 102
effective length of query: 436
effective length of database: 6,144,680
effective search space: 2679080480
effective search space used: 2679080480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149576.2


