

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.1 - phase: 1
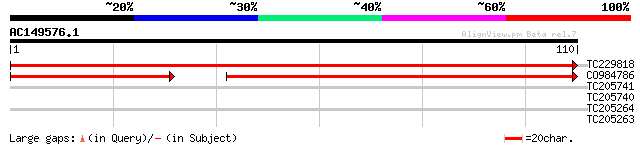
(110 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229818 similar to UP|CYC6_ARATH (Q93VA3) Cytochrome c6, chloro... 207 8e-55
CO984786 130 6e-44
TC205741 homologue to UP|Q852R1 (Q852R1) Serine palmitoyltransfe... 28 0.51
TC205740 homologue to UP|Q852R1 (Q852R1) Serine palmitoyltransfe... 28 0.51
TC205264 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {A... 27 1.9
TC205263 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {A... 24 9.6
>TC229818 similar to UP|CYC6_ARATH (Q93VA3) Cytochrome c6, chloroplast
precursor (Soluble cytochrome f) (Cytochrome c553)
(Cytochrome c-553) (Cytochrome c-552) (Atc6), partial
(71%)
Length = 786
Score = 207 bits (526), Expect = 8e-55
Identities = 96/110 (87%), Positives = 103/110 (93%)
Frame = +2
Query: 1 SIAQTIDIQRGATLFSQTCIGCHDAGGNIIQPGSTLFTKDLQRNGVDTEEEIYRVTYYGK 60
S AQT+DIQRG TLF Q CIGCHDAGGNIIQPG+TLF KDLQRNGVDTEE IY VTYYGK
Sbjct: 278 SSAQTVDIQRGTTLFRQACIGCHDAGGNIIQPGATLFAKDLQRNGVDTEEAIYGVTYYGK 457
Query: 61 GRMPGFGKECMPRGQCTFGARLEDEDIKILAEFVKLQADKGWPSIETEQK 110
GRMPGFGKECMPRGQCTFGARLEDEDI+ILAEFVKLQAD+GWPSIET+++
Sbjct: 458 GRMPGFGKECMPRGQCTFGARLEDEDIQILAEFVKLQADQGWPSIETKEE 607
>CO984786
Length = 686
Score = 130 bits (328), Expect(2) = 6e-44
Identities = 60/68 (88%), Positives = 64/68 (93%)
Frame = -3
Query: 43 RNGVDTEEEIYRVTYYGKGRMPGFGKECMPRGQCTFGARLEDEDIKILAEFVKLQADKGW 102
RN VDTEE IYRVTYYGK RMPGFGKECMPRGQCTFGARLEDEDIK LAEFVKLQAD+GW
Sbjct: 294 RNEVDTEEAIYRVTYYGKARMPGFGKECMPRGQCTFGARLEDEDIKTLAEFVKLQADQGW 115
Query: 103 PSIETEQK 110
PSIET+++
Sbjct: 114 PSIETKEE 91
Score = 61.6 bits (148), Expect(2) = 6e-44
Identities = 27/32 (84%), Positives = 28/32 (87%)
Frame = -2
Query: 1 SIAQTIDIQRGATLFSQTCIGCHDAGGNIIQP 32
S AQT+DIQRG TLF Q CIGCHDAGGNIIQP
Sbjct: 388 SSAQTVDIQRGTTLFRQACIGCHDAGGNIIQP 293
>TC205741 homologue to UP|Q852R1 (Q852R1) Serine palmitoyltransferase, complete
Length = 2191
Score = 28.5 bits (62), Expect = 0.51
Identities = 12/46 (26%), Positives = 22/46 (47%), Gaps = 4/46 (8%)
Frame = +3
Query: 30 IQPGSTLFTKDLQRNGV----DTEEEIYRVTYYGKGRMPGFGKECM 71
I+ S F +LQ+ G D + + + Y ++P F +EC+
Sbjct: 1281 IRENSNFFRSELQKMGFEVLGDNDSPVMPIMLYNPAKIPAFSRECL 1418
>TC205740 homologue to UP|Q852R1 (Q852R1) Serine palmitoyltransferase,
partial (24%)
Length = 911
Score = 28.5 bits (62), Expect = 0.51
Identities = 12/46 (26%), Positives = 22/46 (47%), Gaps = 4/46 (8%)
Frame = +3
Query: 30 IQPGSTLFTKDLQRNGV----DTEEEIYRVTYYGKGRMPGFGKECM 71
I+ S F +LQ+ G D + + + Y ++P F +EC+
Sbjct: 30 IRENSNFFRSELQKMGFEVLGDNDSPVMPIMLYNPAKIPAFSRECL 167
>TC205264 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {Arabidopsis
thaliana;} , partial (62%)
Length = 728
Score = 26.6 bits (57), Expect = 1.9
Identities = 15/38 (39%), Positives = 20/38 (52%)
Frame = +2
Query: 73 RGQCTFGARLEDEDIKILAEFVKLQADKGWPSIETEQK 110
R +FG+RLED K +AE + K W S +E K
Sbjct: 155 RATSSFGSRLEDTIKKTVAENPVVLYSKTWCSYSSEVK 268
>TC205263 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {Arabidopsis
thaliana;} , partial (64%)
Length = 966
Score = 24.3 bits (51), Expect = 9.6
Identities = 13/34 (38%), Positives = 19/34 (55%)
Frame = +1
Query: 77 TFGARLEDEDIKILAEFVKLQADKGWPSIETEQK 110
+FG+RLED K +AE + K W + +E K
Sbjct: 202 SFGSRLEDTIKKTVAENPVVVYSKTWCTYSSEVK 303
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,421,847
Number of Sequences: 63676
Number of extensions: 47330
Number of successful extensions: 174
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 174
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 174
length of query: 110
length of database: 12,639,632
effective HSP length: 86
effective length of query: 24
effective length of database: 7,163,496
effective search space: 171923904
effective search space used: 171923904
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC149576.1


